
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,236,408 – 4,236,508 |
| Length | 100 |
| Max. P | 0.818413 |

| Location | 4,236,408 – 4,236,508 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
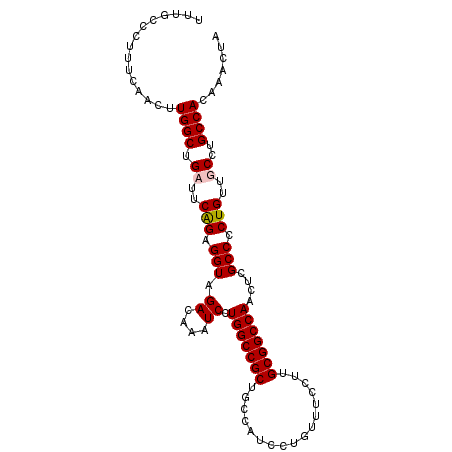
>2L_DroMel_CAF1 4236408 100 + 22407834 UAGUUUGUGGCAGGCAACAGAGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCUCUGAGGCAGCCAAGUUGAAAGGGCAAA ..((((.((((..((..((((((((((((((((((..(....).(.....).)))))))..)))))....))))))..)).))))........))))... ( -35.30) >DroSec_CAF1 11542 100 + 1 UAGUUUGUGGCAGGCAACAGGGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCUCUGGAGCAGCCAUGUUGAAAGGGCAAA ......(((((..((..((((((((((((((((((..(....).(.....).)))))))..)))))....))))))..)).))))).............. ( -35.70) >DroEre_CAF1 11917 100 + 1 UAGUUUGUGGCAGGCAACAGGGGCAAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCUCUGAAUCAGCCAACUUGAAAGGGCCAA .......((((.(((..((((((((((((((((((..(....).(.....).)))))))..)))))....)))))).....)))..((.....)))))). ( -33.50) >DroYak_CAF1 19908 100 + 1 UAGUUUGUGGCAGGCAACAGGGGCGAAUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCUCCGAAUCAGCCAAGUUGAAGGGGCAAA .......((.(...((((...(((...((((((((..(....).(.....).))))))))((((((........)))))).)))..))))....).)).. ( -32.20) >consensus UAGUUUGUGGCAGGCAACAGGGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCUCUGAAGCAGCCAAGUUGAAAGGGCAAA .......((((.(....)..(((((((((((((((..(....).(.....).)))))))..))))))))............))))............... (-30.59 = -30.02 + -0.56)



| Location | 4,236,408 – 4,236,508 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.16 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4236408 100 - 22407834 UUUGCCCUUUCAACUUGGCUGCCUCAGAGGUAGACAAAUCCUGGCCGCUGCCAUCCUGUUUCCUUGCGGCCAACUCGCCUCUGUUGCCUGCCACAAACUA ...............((((.((..(((((((.((....)).(((((((.................)))))))....)))))))..))..))))....... ( -32.43) >DroSec_CAF1 11542 100 - 1 UUUGCCCUUUCAACAUGGCUGCUCCAGAGGUAGACAAAUCCUGGCCGCUGCCAUCCUGUUUCCUUGCGGCCAACUCGCCCCUGUUGCCUGCCACAAACUA ...............((((.((..(((.(((.((....)).(((((((.................)))))))....))).)))..))..))))....... ( -27.83) >DroEre_CAF1 11917 100 - 1 UUGGCCCUUUCAAGUUGGCUGAUUCAGAGGUAGACAAAUCCUGGCCGCUGCCAUCCUGUUUCCUUGCGGCCAACUUGCCCCUGUUGCCUGCCACAAACUA ..(((.....(((((((((((.....((((.(((((.....((((....))))...))))))))).)))))))))))........)))............ ( -32.42) >DroYak_CAF1 19908 100 - 1 UUUGCCCCUUCAACUUGGCUGAUUCGGAGGUAGACAAAUCCUGGCCGCUGCCAUCCUGUUUCCUUGCGGCCAAUUCGCCCCUGUUGCCUGCCACAAACUA ...((.....((((..(((......(((.(....)...)))(((((((.................)))))))....)))...))))...))......... ( -23.63) >consensus UUUGCCCUUUCAACUUGGCUGAUUCAGAGGUAGACAAAUCCUGGCCGCUGCCAUCCUGUUUCCUUGCGGCCAACUCGCCCCUGUUGCCUGCCACAAACUA ...............((((.((..(((.(((.((....)).(((((((.................)))))))....))).)))..))..))))....... (-23.84 = -24.16 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:42 2006