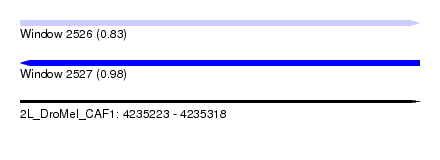
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,235,223 – 4,235,318 |
| Length | 95 |
| Max. P | 0.981475 |

| Location | 4,235,223 – 4,235,318 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -24.42 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
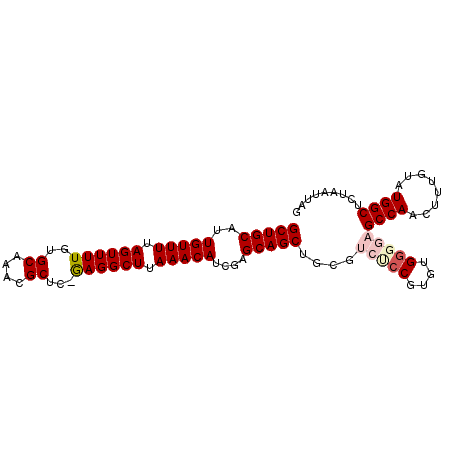
>2L_DroMel_CAF1 4235223 95 + 22407834 GCUGCAUUGUUUUAGUUUUGUGCAAACGCUC-GAGGCUUAAACAUCGAGCAGCUGCGUCUCCGUGCGAGGAGCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.(((((((.((....)).)-)))))).)))))....)))))((((......)))).(((((((........)))))))...... ( -33.70) >DroSec_CAF1 10340 95 + 1 GCUGCAUUGUUUUAGUUUUGUGCAAACGCCG-GAGGCUUAAACAUCGAGCAGCUGCGUCCCCAUGUGGGGAGCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.((((((..((....))..-)))))).)))))....)))))....(((((....)))))((((........))))......... ( -31.70) >DroSim_CAF1 17361 95 + 1 GCUGCAUUGUUUUAGUUUUGUGCAAACGCCG-GAGGCUUAAACAUCGAGCAGCUGCGUCCCCGUGUGGGGAGCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.((((((..((....))..-)))))).)))))....)))))....(((((....)))))((((........))))......... ( -31.00) >DroEre_CAF1 10650 92 + 1 GCUGCAUUGUUUUAGUUUUGUGCAAACGCUC-GAGGCUUAAACAUCGAGCAGCUGCGUCUCCGUGUGG---GCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.(((((((.((....)).)-)))))).)))))....))))).(((....)))..((---((((........))))))....... ( -30.50) >DroYak_CAF1 18638 93 + 1 GCUGCAUUGUUUUAGUUUUGUGCAAACGCUCCAAGGCUUAAACAUCGAGCAGCUGCGUCUCCGUGUGG---GCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.(((((((.((....))..))))))).)))))....))))).(((....)))..((---((((........))))))....... ( -28.80) >consensus GCUGCAUUGUUUUAGUUUUGUGCAAACGCUC_GAGGCUUAAACAUCGAGCAGCUGCGUCUCCGUGUGGGGAGCCAACUUUGUAUGGCUCUAAUUAG (((((..(((((.((((((..((....))...)))))).)))))....)))))....(((((....)))))((((........))))......... (-24.42 = -25.58 + 1.16)

| Location | 4,235,223 – 4,235,318 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
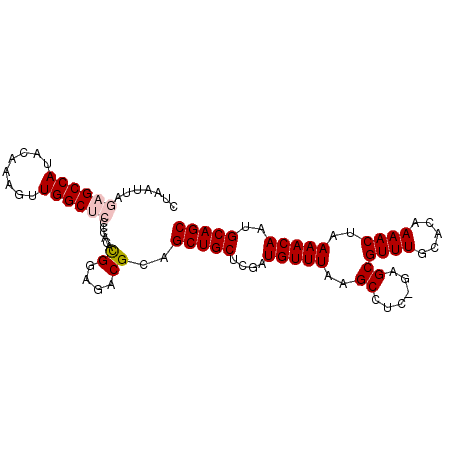
>2L_DroMel_CAF1 4235223 95 - 22407834 CUAAUUAGAGCCAUACAAAGUUGGCUCCUCGCACGGAGACGCAGCUGCUCGAUGUUUAAGCCUC-GAGCGUUUGCACAAAACUAAAACAAUGCAGC .......((((((........))))))...(((.(....)((((.(((((((.((....)).))-))))).))))...............)))... ( -31.70) >DroSec_CAF1 10340 95 - 1 CUAAUUAGAGCCAUACAAAGUUGGCUCCCCACAUGGGGACGCAGCUGCUCGAUGUUUAAGCCUC-CGGCGUUUGCACAAAACUAAAACAAUGCAGC .........((((........))))(((((....)))))....(((((....(((((..(((..-.)))((((.....))))..)))))..))))) ( -28.70) >DroSim_CAF1 17361 95 - 1 CUAAUUAGAGCCAUACAAAGUUGGCUCCCCACACGGGGACGCAGCUGCUCGAUGUUUAAGCCUC-CGGCGUUUGCACAAAACUAAAACAAUGCAGC .........((((........))))(((((....)))))....(((((....(((((..(((..-.)))((((.....))))..)))))..))))) ( -28.40) >DroEre_CAF1 10650 92 - 1 CUAAUUAGAGCCAUACAAAGUUGGC---CCACACGGAGACGCAGCUGCUCGAUGUUUAAGCCUC-GAGCGUUUGCACAAAACUAAAACAAUGCAGC ....((((.((((........))))---......(....)((((.(((((((.((....)).))-))))).))))......))))........... ( -25.50) >DroYak_CAF1 18638 93 - 1 CUAAUUAGAGCCAUACAAAGUUGGC---CCACACGGAGACGCAGCUGCUCGAUGUUUAAGCCUUGGAGCGUUUGCACAAAACUAAAACAAUGCAGC .......(.((((........))))---.)...((....))..(((((....(((((.((..(((..((....)).)))..)).)))))..))))) ( -23.10) >consensus CUAAUUAGAGCCAUACAAAGUUGGCUCCCCACACGGAGACGCAGCUGCUCGAUGUUUAAGCCUC_GAGCGUUUGCACAAAACUAAAACAAUGCAGC .......((((((........))))))......((....))..(((((....(((((..((......))((((.....))))..)))))..))))) (-22.70 = -23.34 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:40 2006