
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,186,306 – 4,186,418 |
| Length | 112 |
| Max. P | 0.817747 |

| Location | 4,186,306 – 4,186,418 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.88 |
| Mean single sequence MFE | -47.82 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.38 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4186306 112 - 22407834 UAGUGGCGGCAUCGGCGGCUGCUCC--UCGGUUGGCUAGCCUACGGCAUCAUCCUCGGUUGGCUGCUCUGGGAAAUGCUGCUGCGGCCCUCAGUUACAGGCCCAA------AAUCUAAUC ..((((((((..(((((((...(((--.(((..((((((((...((......))..))))))))...))))))...)))))))..)))....)))))........------......... ( -39.30) >DroPse_CAF1 20922 111 - 1 UAGUGGCGGCGUCGGCGGCUGCUCUGGACCGUUG--UGGCCUAUGCCCUACUGCUGGGCUGGCUGCUGGUGGAGAUGCUGCUGCCGGUGCC-GCUGGUGCCUCCG------AAUGGCACG ....((((((..(((((((.(((((..((((..(--..(((...((((.......)))).)))..)))))..))).)).)))))))..)))-))).(((((....------...))))). ( -66.00) >DroSim_CAF1 730 112 - 1 UAGUGGCGGCAUCGGCGGCUGCUCC--UCGGUUGGCUGGCCUACGGCAUCAUCCUCGGUUGGCUGCUCUGGGAACUGCUGCUGCGGCCCUCAGUUACUGGCCCAA------AAUCUAAUC ..(.((((((..(((((((.(.(((--.(((..(((..(((...((......))..)))..)))...)))))).).)))))))..)))..(((...)))))))..------......... ( -41.30) >DroWil_CAF1 8504 109 - 1 UAGUGGCGACAUCGGCGGUUAUUGU--UCUCAUUAUUGGCCUAUGCCCUUGUGAUGGCUUGGCUACUUUUGGAAAUUCUAAUGCCGCCGU-------UGUCUCAGUAUAGCAAUUUAA-- ((.(((.((((.(((((((....((--(..((..(.(((((...(((........)))..))))).)..))..)))......))))))).-------))))))).))...........-- ( -37.00) >DroYak_CAF1 765 112 - 1 UAGUGGCGGCAUCGGCGGCUGCUCC--UCGGUUGGCUAGCCUAUGGCAUCAUCCUCGGUUGGCUGCUCUGGGAACUGCUGCUGCGGCCCUCAAAUACUGGGCUAA------CAUCUAAUC ..(..((((((..(((....))).(--((((..((((((((...((......))..))))))))...)))))...))))))..)(((((.........)))))..------......... ( -41.40) >DroPer_CAF1 20950 111 - 1 UAGUGGCGGCGUCGGCGGCUGCUCUGGACCGUUG--UGGCCUAUGCCCUACUGCUGGGCUGGCUGCUGGUGGAGAUGCUGCUGCCGGUGCC-GCUGGAGCCUCCG------ACUGGCACG ....((((((..(((((((.(((((..((((..(--..(((...((((.......)))).)))..)))))..))).)).)))))))..)))-)))...(((....------...)))... ( -61.90) >consensus UAGUGGCGGCAUCGGCGGCUGCUCC__UCGGUUGGCUGGCCUAUGCCAUCAUCCUCGGUUGGCUGCUCUGGGAAAUGCUGCUGCCGCCCUC_GCUACUGGCCCAA______AAUCUAAUC ..((((((((((((((((((...(......)......((((...((......))..)))))))))))....))..))))))))).(((..........)))................... (-20.02 = -19.38 + -0.64)

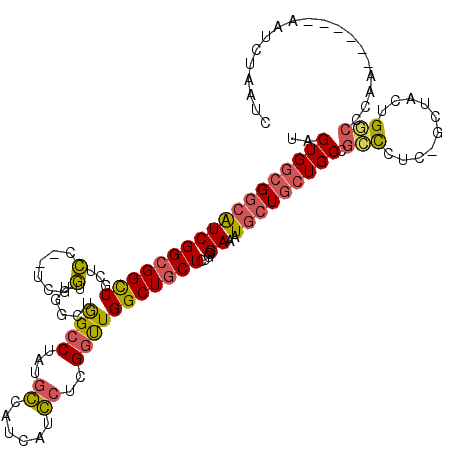
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:27 2006