
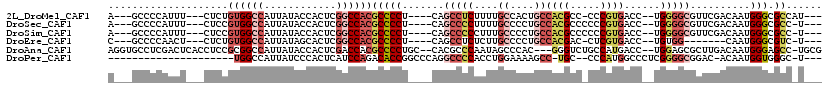
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,168,347 – 4,168,450 |
| Length | 103 |
| Max. P | 0.961706 |

| Location | 4,168,347 – 4,168,450 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.88 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -23.04 |
| Energy contribution | -25.60 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
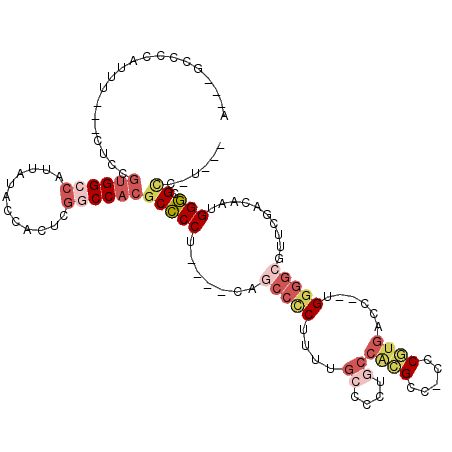
>2L_DroMel_CAF1 4168347 103 + 22407834 ---AUGGCGCCCAUUGUCGAACGCCCCA--GGUCACGGG-GGCGUGGCAGUGGCAAAAGAGGCUG----AGGGGCGUGGCCGAGUGGUAUAAUGGCCACAGAG---AAAUGGGGC---U ---..(((.((((((.(((.(((((((.--.((((((..-..))))))...(((.......))).----.)))))))...)))(((((......)))))....---.))))))))---) ( -46.30) >DroSec_CAF1 3218 103 + 1 ---A-GGCGCCCAUUGUCGAACGCCCCA--GGUCACGGGGGGCGUGGCAGGGGCAAAAGGGGCUG----AGGGGCGUGGCCGAGUGGUAUAAUGGCCACGGAG---AAAUGGGGC---U ---.-(((.((((((.(((.(((((((.--.((((((.....))))))...(((.......))).----.)))))))...)))(((((......)))))....---.))))))))---) ( -46.60) >DroSim_CAF1 3372 103 + 1 ---A-GGCGCCCAUUGUCGAACGCCCCA--GGUCACGGGGGGCGUGGCAGGGGCAAAGGGGGCUG----AGGGGCGUGGCCGAGUGGUAUAAUGGCCACGGAG---AAAUGGGGC---U ---.-(((.((((((.(((.(((((((.--.((((((.....))))))...(((.......))).----.)))))))...)))(((((......)))))....---.))))))))---) ( -46.60) >DroEre_CAF1 3457 95 + 1 ---A-GACGCCCAUUG-------CCACA--GGUCACGAG-GUCGUGGCAGGGGCAAGAGAGGCUG----AGGGGCGUGGCCGAGUGCUAUAAUGGCCACAGAG---AGUUGGGGC---G ---.-..(((((.(((-------((.(.--.((((((..-..))))))..)))))).....(((.----...)))(((((((..........)))))))....---.....))))---) ( -40.20) >DroAna_CAF1 3877 111 + 1 CGCA-GGCUCCCAUUGUCAAGCGCUCCA--GGUCAUGGCAGACCC---GUGGGCUAUUGGGCGUG--GCAGGGGCGUGGUCGAGUGGUAUAAUGGCCGCGGAGGUGAGUCGAGGCACCU .((.-(((((((.(((....(((((((.--.((((((.(((.((.---...))))....).))))--)).)))))))...)))(((((......)))))...)).)))))...)).... ( -44.80) >DroPer_CAF1 69450 90 + 1 ---A-GCCCACCAUUGU-GUCCGCCCCGAGGGCCAUGGG--GCA-GGCUUUUCCAGGUGGGGCCUGGGCCGGUGUCUGGAUGAGUGGGAUAAUGGCCA--------------------- ---.-.....(((((((-.(((((((((.......))))--)).-.((((.(((((..(.(((....)))..)..))))).))))))))))))))...--------------------- ( -38.60) >consensus ___A_GGCGCCCAUUGUCGAACGCCCCA__GGUCACGGG_GGCGUGGCAGGGGCAAAAGGGGCUG____AGGGGCGUGGCCGAGUGGUAUAAUGGCCACGGAG___AAAUGGGGC___U ........((((..........(((((....((((((.....))))))...(((.......)))......)))))(((((((..........)))))))............)))).... (-23.04 = -25.60 + 2.56)



| Location | 4,168,347 – 4,168,450 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.88 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -15.81 |
| Energy contribution | -17.81 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4168347 103 - 22407834 A---GCCCCAUUU---CUCUGUGGCCAUUAUACCACUCGGCCACGCCCCU----CAGCCUCUUUUGCCACUGCCACGCC-CCCGUGACC--UGGGGCGUUCGACAAUGGGCGCCAU--- .---(((((((((---(...((((((............))))))(((((.----...........((....))((((..-..))))...--.)))))....)).)))))).))...--- ( -36.30) >DroSec_CAF1 3218 103 - 1 A---GCCCCAUUU---CUCCGUGGCCAUUAUACCACUCGGCCACGCCCCU----CAGCCCCUUUUGCCCCUGCCACGCCCCCCGUGACC--UGGGGCGUUCGACAAUGGGCGCC-U--- .---(((((((((---(..(((((((............))))))).....----...........(((((.(.((((.....)))).).--.)))))....)).)))))).)).-.--- ( -38.30) >DroSim_CAF1 3372 103 - 1 A---GCCCCAUUU---CUCCGUGGCCAUUAUACCACUCGGCCACGCCCCU----CAGCCCCCUUUGCCCCUGCCACGCCCCCCGUGACC--UGGGGCGUUCGACAAUGGGCGCC-U--- .---(((((((((---(..(((((((............))))))).....----...........(((((.(.((((.....)))).).--.)))))....)).)))))).)).-.--- ( -38.30) >DroEre_CAF1 3457 95 - 1 C---GCCCCAACU---CUCUGUGGCCAUUAUAGCACUCGGCCACGCCCCU----CAGCCUCUCUUGCCCCUGCCACGAC-CUCGUGACC--UGUGG-------CAAUGGGCGUC-U--- (---((((.....---....((((((............))))))(((...----(((........((....))((((..-..))))..)--)).))-------)...)))))..-.--- ( -31.00) >DroAna_CAF1 3877 111 - 1 AGGUGCCUCGACUCACCUCCGCGGCCAUUAUACCACUCGACCACGCCCCUGC--CACGCCCAAUAGCCCAC---GGGUCUGCCAUGACC--UGGAGCGCUUGACAAUGGGAGCC-UGCG (((((........))))).((((((...................((....))--....((((..((((..(---(((((......))))--))..).)))......)))).)))-.))) ( -30.40) >DroPer_CAF1 69450 90 - 1 ---------------------UGGCCAUUAUCCCACUCAUCCAGACACCGGCCCAGGCCCCACCUGGAAAAGCC-UGC--CCCAUGGCCCUCGGGGCGGAC-ACAAUGGUGGGC-U--- ---------------------..........(((((.(((((((.....(((....)))....)))))...(.(-(((--(((.........))))))).)-....))))))).-.--- ( -33.80) >consensus A___GCCCCAUUU___CUCCGUGGCCAUUAUACCACUCGGCCACGCCCCU____CAGCCCCUUUUGCCCCUGCCACGCC_CCCGUGACC__UGGGGCGUUCGACAAUGGGCGCC_U___ ....................((((((............))))))(((((.......(((((....((....))((((.....))))......)))))..........))).))...... (-15.81 = -17.81 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:15 2006