
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,162,753 – 4,162,888 |
| Length | 135 |
| Max. P | 0.955754 |

| Location | 4,162,753 – 4,162,853 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 96.69 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
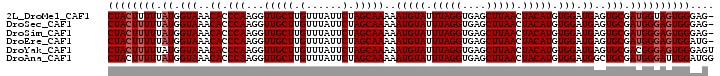
>2L_DroMel_CAF1 4162753 100 - 22407834 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGAUGGUAGUGGGAG- (((((..((((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).)))).)))))...- ( -25.50) >DroSec_CAF1 292927 100 - 1 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGAUGGGAGUGGGAG- ((((((((.((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).))))))))))...- ( -26.20) >DroSim_CAF1 295553 100 - 1 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGAUGGGAGUGGGAG- ((((((((.((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).))))))))))...- ( -26.20) >DroEre_CAF1 310097 100 - 1 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGAUGGGAGUGGAUG- ((((((((.((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).))))))))))...- ( -26.00) >DroYak_CAF1 315259 101 - 1 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGACGGGAGUGGGAGU ((((((((..(.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).).)))))))).... ( -23.90) >DroAna_CAF1 341961 101 - 1 CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGGCUGCGAUGGGAUUGGAUGG (((..((((((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).)))))).))).... ( -23.60) >consensus CUACUUUUUAUGGUAAACACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUACAUGUGGAUGAGUGCGAUGGGAGUGGGAG_ ((((((((.((.(((..((.(((...(((((.(......).)))))..(((((.(((((....))))).))))).))).))..))).)))))))))).... (-24.42 = -24.92 + 0.50)


| Location | 4,162,780 – 4,162,888 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.37 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4162780 108 - 22407834 CG----CAUUUGCUACGCAAUU-UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACA-CCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUAC---A--- .(----(....))......(((-(((((......(((((((..........))))))).-..((((....)))).........))))))))(((.(((((....))))).)))---.--- ( -24.10) >DroVir_CAF1 438877 113 - 1 CUCACUCGCUCCAUUCGCAAUU-UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACAACCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGCGCUUAACUAC---A--- ......(((.((.......(((-(((((......(((((((..........)))))))....((((....)))).........)))))))).......)).))).........---.--- ( -24.94) >DroGri_CAF1 405572 119 - 1 CUCGCACACUCCAUACGCAAUU-UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACAUUCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGCGCUUAACUACUACGUGA ..(((((.((..((((...(((-(((((......(((((((..........)))))))....((((....)))).........))))))))))))..)))))))................ ( -28.10) >DroYak_CAF1 315287 108 - 1 CG----CAUUUGCUACGCAAUU-UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACA-CCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUAC---A--- .(----(....))......(((-(((((......(((((((..........))))))).-..((((....)))).........))))))))(((.(((((....))))).)))---.--- ( -24.10) >DroMoj_CAF1 472853 114 - 1 CUUACUCGCUCCAUACGCAAUUUUUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACAUACCAAGGUUGCUUGUUUAUUCUAGCAAAAGUGUAUUUAGGUGCGCUUAGCUAU---G--- .......((.((....((((....))))......(((((((..........))))))).......))..))..........((((..((((((((....)))))))).)))).---.--- ( -27.00) >DroAna_CAF1 341989 108 - 1 CG----CAUUUGCUACGCAAUU-UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACA-CCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUAC---A--- .(----(....))......(((-(((((......(((((((..........))))))).-..((((....)))).........))))))))(((.(((((....))))).)))---.--- ( -24.10) >consensus CG____CACUCCAUACGCAAUU_UUUGCACUCAAGUUUACCUACUUUUUAUGGUAAACA_CCCAAGGUUGCUUGUUUAUUCUAGCAAAAAUGUAUUUAGGUGAGCUUAACUAC___A___ ................((((....))))......(((((((..........))))))).........(((((.(......).)))))....(((.(((((....))))).)))....... (-19.64 = -19.37 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:10 2006