| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,162,408 – 4,162,515 |
| Length | 107 |
| Max. P | 0.693720 |

| Location | 4,162,408 – 4,162,515 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -6.96 |
| Energy contribution | -6.96 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4162408 107 + 22407834 UUUUUAUUGGUCGCUGCAUUAUGCAUGUUUGUCACACAUUUAAUUAUUCAC--ACUCGCACAU--------ACGCCAUUUCUUCCCACCUCCUGUAUGCGUGUGUGUGUGUGUGUAU .......((((.((.((((.....))))..)).)).))..........(((--((.(((((((--------((((......................))))))))))).)))))... ( -26.85) >DroSec_CAF1 292586 103 + 1 U-UUUAUUUGUCGCUGCAUUAUGCAUGUUUGUCACACAUUUAAUUAUUCAC--ACUCGCACAU--------AC-CUAUUUCUUGCCACCUCCUGUAUGCGAGUG--UGUGUGUGUAU .-..........(((((.....))).))....((((((..........(((--(((((((.((--------(.-..................))).))))))))--))))))))... ( -23.11) >DroSim_CAF1 295221 94 + 1 U-UUUAUUUGUCGCUGCAUUAUGCAUGUUUGUCACACAUUUAAUUAUUCAC--ACUCGCACAU--------AC-CCAUUUCU-------UCCUGUAUGCG--UG--UGUGUGUGUAU .-......(((.((.((((.....))))..)).)))........(((.(((--((.(((.(((--------((-........-------....))))).)--))--.))))).))). ( -19.40) >DroEre_CAF1 309755 107 + 1 U-UUCAUUUGUCGCUGCAUUAUGCAUAUUUGUCACACAUUUAAUUAUUCACAUACUCGCACAUAUGUGUGUAC-CUCUUUUCUGCCACCCC------GCUCAUG--UGUGUGUGUAU .-..((..(((.((........)))))..))....................((((.((((((((((.(((...-................)------)).))))--)))))).)))) ( -21.31) >DroYak_CAF1 314915 92 + 1 U-UUUAUUCGCCGCUGCAUUAUGCAUGUUUGUCACACAUUUAAUUAUUCAC--ACUCGCACAU--------AC-CUAUUUUCUGUCCCC---------AUC--G--UGUGUGUGUGU .-..........(((((.....))).))....................(((--((.(((((..--------..-...............---------...--)--)))).))))). ( -14.35) >consensus U_UUUAUUUGUCGCUGCAUUAUGCAUGUUUGUCACACAUUUAAUUAUUCAC__ACUCGCACAU________AC_CUAUUUCUUGCCACCUCCUGUAUGCGC_UG__UGUGUGUGUAU ..............(((.....)))............................((.((((((............................................)))))).)).. ( -6.96 = -6.96 + -0.00)

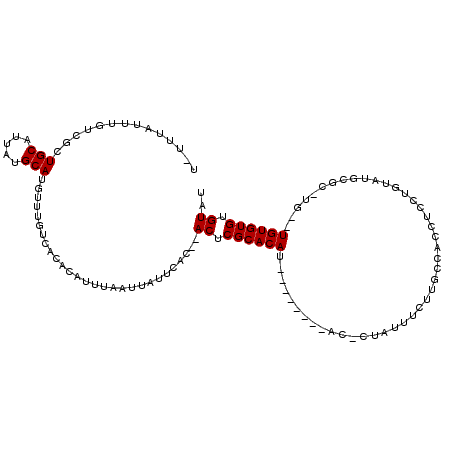
| Location | 4,162,408 – 4,162,515 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -6.10 |
| Energy contribution | -6.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4162408 107 - 22407834 AUACACACACACACACACGCAUACAGGAGGUGGGAAGAAAUGGCGU--------AUGUGCGAGU--GUGAAUAAUUAAAUGUGUGACAAACAUGCAUAAUGCAGCGACCAAUAAAAA .....(((((((((((.((((((.(.(..((........))..).)--------.)))))).))--)))..........)))))).......(((.....))).............. ( -24.10) >DroSec_CAF1 292586 103 - 1 AUACACACACA--CACUCGCAUACAGGAGGUGGCAAGAAAUAG-GU--------AUGUGCGAGU--GUGAAUAAUUAAAUGUGUGACAAACAUGCAUAAUGCAGCGACAAAUAAA-A ...((((((((--((((((((((......((........))..-..--------.)))))))))--)))..........)))))).......(((.....)))............-. ( -26.40) >DroSim_CAF1 295221 94 - 1 AUACACACACA--CA--CGCAUACAGGA-------AGAAAUGG-GU--------AUGUGCGAGU--GUGAAUAAUUAAAUGUGUGACAAACAUGCAUAAUGCAGCGACAAAUAAA-A .....(((((.--(.--(((((((....-------(....)..-))--------))))).).))--))).........(((((((.....)))))))..................-. ( -21.70) >DroEre_CAF1 309755 107 - 1 AUACACACACA--CAUGAGC------GGGGUGGCAGAAAAGAG-GUACACACAUAUGUGCGAGUAUGUGAAUAAUUAAAUGUGUGACAAAUAUGCAUAAUGCAGCGACAAAUGAA-A ...((((((((--((((..(------(..(((.(........)-.))).(((....)))))..))))))..........)))))).......(((.....)))............-. ( -20.70) >DroYak_CAF1 314915 92 - 1 ACACACACACA--C--GAU---------GGGGACAGAAAAUAG-GU--------AUGUGCGAGU--GUGAAUAAUUAAAUGUGUGACAAACAUGCAUAAUGCAGCGGCGAAUAAA-A .(((((.(.((--(--(.(---------(....))........-..--------.)))).).))--))).........(((((((.....)))))))..(((....)))......-. ( -17.90) >consensus AUACACACACA__CA_GCGCAUACAGGAGGUGGCAAGAAAUAG_GU________AUGUGCGAGU__GUGAAUAAUUAAAUGUGUGACAAACAUGCAUAAUGCAGCGACAAAUAAA_A ...((((((......................................................................)))))).......(((.....))).............. ( -6.10 = -6.10 + 0.00)

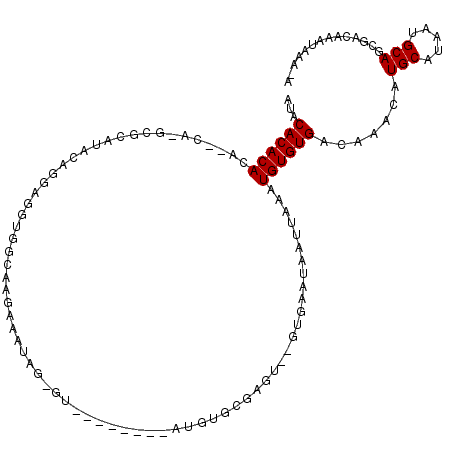
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:08 2006