
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,156,127 – 4,156,225 |
| Length | 98 |
| Max. P | 0.967844 |

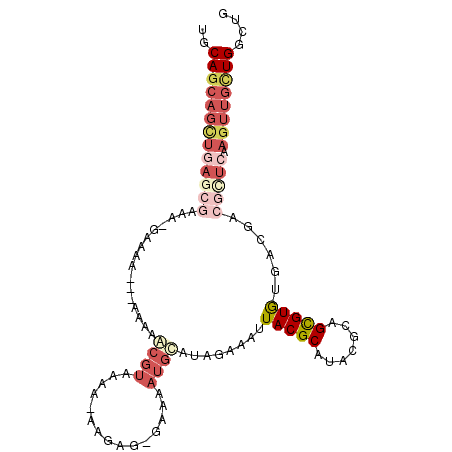
| Location | 4,156,127 – 4,156,225 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -12.60 |
| Energy contribution | -15.16 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
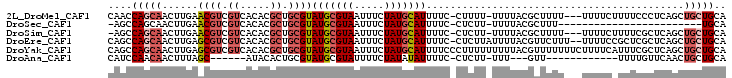
>2L_DroMel_CAF1 4156127 98 + 22407834 UGCAGCAGCUGAGGGAAAAGAAAA---AAAAGCGUAAAA-AAAAG-GAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGUUCAAGUUGCUGGUUG ..(((((((((((.(.........---....((((....-.....-....))))..........((((((((...)))))).)).).))).)))))))).... ( -24.52) >DroSec_CAF1 284849 76 + 1 UGCA------------------------AAAGCGUAAAA-AAGAG-GAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGUUCAAGUUGCUGGCU- .((.------------------------...((((((..-.....-(......).......))))))....(((((...(((......))).)))))..)).- ( -15.44) >DroSim_CAF1 289031 97 + 1 UGCAGCAGCUGAGCGAAAAGAAAA---AAAAGCGUAAAA-AAGAG-GAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGUUCAAGUUGCUGGCU- ..(((((((((((((.........---....((((....-.....-....))))..........((((((((...)))))).)).))))).))))))))...- ( -28.42) >DroEre_CAF1 300956 100 + 1 UGCAGCAGCUGAGCGAGCGGAAAA--AAAGAACGUAAAAUAAGAG-GAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGCUCAAGUUGCUGGCUG ..(((((((((((((.(((.....--......)))..........-..................((((((((...)))))).)).))))).)))))))).... ( -30.20) >DroYak_CAF1 308055 103 + 1 UGCAGCAGCUGAGCGAAAUGAAAAGAAAAAAACGUAAAAAAAAAGGGAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGCUCAAGUUGCUGGCUG ..(((((((((((((..................(((...............)))..........((((((((...)))))).)).))))).)))))))).... ( -27.86) >DroAna_CAF1 334736 80 + 1 UGCAGCAGUUGAACAAAA------------AAC---AAA-AAGAG-GAAAAUAUAUAGAAAAUACGCAUACGCAGUGUAU------GCUAAAGUUGUUGGAUG .(((((.(((........------------)))---...-.....-...................((((((.....))))------))....)))))...... ( -12.70) >consensus UGCAGCAGCUGAGCGAAA_GAAAA___AAAAACGUAAAA_AAGAG_GAAAAUGCAUAGAAAUUACGCAUACGCAGCGUGUGACGACGCUCAAGUUGCUGGCUG ..(((((((((((((................((((...............))))........(((((.......)))))......))))).)))))))).... (-12.60 = -15.16 + 2.56)


| Location | 4,156,127 – 4,156,225 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -9.44 |
| Energy contribution | -10.55 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4156127 98 - 22407834 CAACCAGCAACUUGAACGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUC-CUUUU-UUUUACGCUUUU---UUUUCUUUUCCCUCAGCUGCUGCA ....(((((.((.((.(((......)))..(((((((((((.....)))))).....-.....-...)))))....---............)))).))))).. ( -16.91) >DroSec_CAF1 284849 76 - 1 -AGCCAGCAACUUGAACGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUC-CUCUU-UUUUACGCUUU------------------------UGCA -.((.(((.......((((.((.....)).))))(((((((.....)))))))....-.....-......)))..------------------------.)). ( -13.70) >DroSim_CAF1 289031 97 - 1 -AGCCAGCAACUUGAACGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUC-CUCUU-UUUUACGCUUUU---UUUUCUUUUCGCUCAGCUGCUGCA -.((.((((.((.((.((............(((((((((((.....)))))).....-.....-...)))))....---.........)).)))).)))))). ( -19.22) >DroEre_CAF1 300956 100 - 1 CAGCCAGCAACUUGAGCGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUC-CUCUUAUUUUACGUUCUUU--UUUUCCGCUCGCUCAGCUGCUGCA ..((.((((.((.(((((.((.........(((((((((((.....)))))).....-.........))))).....--.....))..))))))).)))))). ( -24.84) >DroYak_CAF1 308055 103 - 1 CAGCCAGCAACUUGAGCGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUCCCUUUUUUUUUACGUUUUUUUCUUUUCAUUUCGCUCAGCUGCUGCA ..((.((((.((.(((((.((.....))..(((((((((((.....))))))...............)))))................))))))).)))))). ( -22.46) >DroAna_CAF1 334736 80 - 1 CAUCCAACAACUUUAGC------AUACACUGCGUAUGCGUAUUUUCUAUAUAUUUUC-CUCUU-UUU---GUU------------UUUUGUUCAACUGCUGCA ((...(((((.....((------((((.....))))))((((.....))))......-.....-.))---)))------------...))............. ( -9.10) >consensus CAGCCAGCAACUUGAACGUCGUCACACGCUGCGUAUGCGUAAUUUCUAUGCAUUUUC_CUCUU_UUUUACGCUUUU___UUUUC_UUUCGCUCAGCUGCUGCA ....(((((......((((.((.....)).))))(((((((.....)))))))...........................................))))).. ( -9.44 = -10.55 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:02 2006