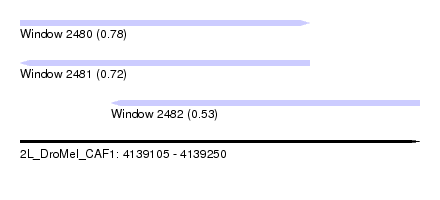
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,139,105 – 4,139,250 |
| Length | 145 |
| Max. P | 0.775881 |

| Location | 4,139,105 – 4,139,210 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
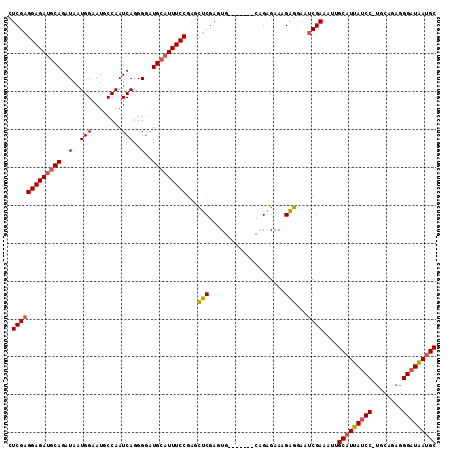
>2L_DroMel_CAF1 4139105 105 + 22407834 CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGGAUGCAUUUCCGAGCUCGAGUG-------CAGAGAAAGAGGAAUCGAAAUUGCAUUAUCC-UGCAGAGGGAUAAUGC .(((((((((((((..(..(((........)))..)..)))))))))...(((.....-------..))).........))))....(((((((((-(.....)))))))))) ( -33.80) >DroSec_CAF1 268558 105 + 1 CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGGAUGCAUUUCCGAGCUCGAGUG-------CGGAGAAAGAGGAAUCGAAAUUGCAUUAUCC-UGCAGAGGGAUAAUGC .(((((((((((((..(..(((........)))..)..)))))))))..((......)-------).............))))....(((((((((-(.....)))))))))) ( -33.60) >DroSim_CAF1 272198 105 + 1 CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGGAUGCAUUUCCGAGCUCGAGUG-------CGGAGAAAGAGGAAUCGAAAUUGCAUUAUCC-UGCAGAGGGAUAAUGC .(((((((((((((..(..(((........)))..)..)))))))))..((......)-------).............))))....(((((((((-(.....)))))))))) ( -33.60) >DroEre_CAF1 282543 105 + 1 CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGAAUGCCUUUCCGAGCUCGAAUG-------CAGAGAAAGAGGAAUCGAAAUUGCAUUAUCC-UGCAGAGGGAUAAUGC .(((((((((.(((.....(((........))).....))).)))))...(((.....-------..))).........))))....(((((((((-(.....)))))))))) ( -28.60) >DroYak_CAF1 287133 105 + 1 CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGGAUGCAUUUCCGAGCUCGAGUG-------CAGAGAAAGAGGAAUCGAAAUUGCAUUAUCC-UGCAGAGGGAUAAUGC .(((((((((((((..(..(((........)))..)..)))))))))...(((.....-------..))).........))))....(((((((((-(.....)))))))))) ( -33.80) >DroAna_CAF1 316205 113 + 1 CUCGCGGAGAUUCAGAUAAUGGGAUGCCAAUCAGGGGUUGCAUUUCCCGAUCCGUAUGCCUUAUCCGGUGAAAGGAAAAUCGAAACUGCAUUACCCCAGCAACGGGAUGAAGC ...((((.......(((((.((.((((..(((.((((.......)))))))..)))).)))))))((((.........))))...)))).(((.(((......))).)))... ( -29.60) >consensus CUCGAGGAGAUGCAGAUAAUGGAAUGCCAAUCAGGGGAUGCAUUUCCGAGCUCGAGUG_______CAGAGAAAGAGGAAUCGAAAUUGCAUUAUCC_UGCAGAGGGAUAAUGC .(((((((((((((..(..(((........)))..)..)))))))))...(((....................)))...))))....(((((((((........))))))))) (-26.59 = -26.73 + 0.14)

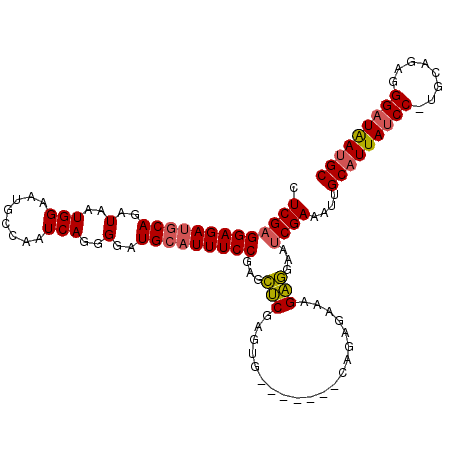
| Location | 4,139,105 – 4,139,210 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4139105 105 - 22407834 GCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CACUCGAGCUCGGAAAUGCAUCCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((.......-)))))))))......................-------..(((((....(((.(((((......((((((....)))..)))))))).)))))))) ( -25.90) >DroSec_CAF1 268558 105 - 1 GCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG-------CACUCGAGCUCGGAAAUGCAUCCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((.......-)))))))))....((((.............(-------(......))..(((.(((((......((((((....)))..)))))))).))))))). ( -26.00) >DroSim_CAF1 272198 105 - 1 GCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG-------CACUCGAGCUCGGAAAUGCAUCCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((.......-)))))))))....((((.............(-------(......))..(((.(((((......((((((....)))..)))))))).))))))). ( -26.00) >DroEre_CAF1 282543 105 - 1 GCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CAUUCGAGCUCGGAAAGGCAUUCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((.......-)))))))))....((((.....((((((..(-------(......))..))))))(((......((((((....)))..))))))......)))). ( -26.10) >DroYak_CAF1 287133 105 - 1 GCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CACUCGAGCUCGGAAAUGCAUCCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((.......-)))))))))......................-------..(((((....(((.(((((......((((((....)))..)))))))).)))))))) ( -25.90) >DroAna_CAF1 316205 113 - 1 GCUUCAUCCCGUUGCUGGGGUAAUGCAGUUUCGAUUUUCCUUUCACCGGAUAAGGCAUACGGAUCGGGAAAUGCAACCCCUGAUUGGCAUCCCAUUAUCUGAAUCUCCGCGAG ((...((((((....))))))...))....((((........))..(((((((((.((.(.(((((((..........))))))).).)).)).))))))).........)). ( -27.60) >consensus GCAUUAUCCCUCUGCA_GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG_______CACUCGAGCUCGGAAAUGCAUCCCCUGAUUGGCAUUCCAUUAUCUGCAUCUCCUCGAG (((((((((........)))))))))....((((...(((....................)))...(((.(((((......((((((....)))..)))))))).))))))). (-21.98 = -22.12 + 0.14)

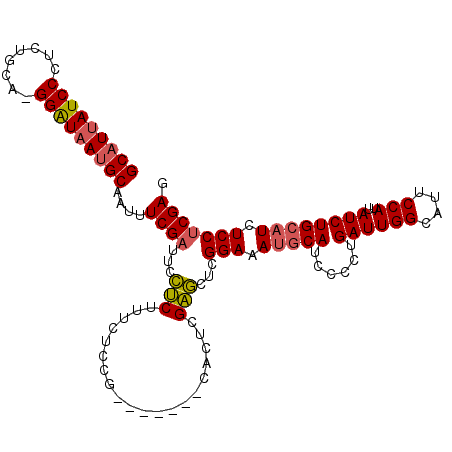
| Location | 4,139,138 – 4,139,250 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.15 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4139138 112 - 22407834 UUACAAAUAAAUAUAUCGAACAUCUUGCUUCGACUGAGGAGCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CACUCGAGCUCGGAAAUGCAUCCCC ...............((((......(((...((..((((((((((((((.......-))))))))).........)))))..))...)-------)).))))((........))...... ( -24.70) >DroSec_CAF1 268591 112 - 1 UUACAAAUAAAUAUGUCGAACAUCUCCCUUCGACUGGAGAGCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG-------CACUCGAGCUCGGAAAUGCAUCCCC ..............((((((........)))))).((((((((((((((.......-)))))))))......((....))..)))))(-------((.(((....)))...)))...... ( -28.90) >DroSim_CAF1 272231 112 - 1 CUACAAAUAAAUAUAUCGAACAUCUCGCUUCGACUGGAGAGCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG-------CACUCGAGCUCGGAAAUGCAUCCCC ..........................(((.(((.(((((((((((((((.......-)))))))))......((....))..))))))-------...))))))..(((......))).. ( -26.70) >DroEre_CAF1 282576 112 - 1 UUACAAAUAAAUAUAUUGAACAUCUCGGUACGACUGAAGAGCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CAUUCGAGCUCGGAAAGGCAUUCCC ..............((((((..((((((.....))).)))(((((((((.......-)))))))))...))))))...(((((((..(-------(......))..)))))))....... ( -26.90) >DroYak_CAF1 287166 112 - 1 UUACAAAUAAAUAUAUUGAACAUCUCGUUUCGACUGAAGAGCAUUAUCCCUCUGCA-GGAUAAUGCAAUUUCGAUUCCUCUUUCUCUG-------CACUCGAGCUCGGAAAUGCAUCCCC ......................((.((.(((((.((.((((((((((((.......-)))))))))......((........))))).-------)).)))))..))))........... ( -22.20) >DroAna_CAF1 316238 119 - 1 AUAUACAUAUGUGUAUUGAAAAUGGCACU-CAAGGUAAAAGCUUCAUCCCGUUGCUGGGGUAAUGCAGUUUCGAUUUUCCUUUCACCGGAUAAGGCAUACGGAUCGGGAAAUGCAACCCC ((((((....))))))....(((((....-..((((....))))....)))))...(((((..((((.(..((((((.((((((....)).)))).....))))))..)..))))))))) ( -27.00) >consensus UUACAAAUAAAUAUAUCGAACAUCUCGCUUCGACUGAAGAGCAUUAUCCCUCUGCA_GGAUAAUGCAAUUUCGAUUCCUCUUUCUCCG_______CACUCGAGCUCGGAAAUGCAUCCCC ...............(((((........))))).......(((((((((........)))))))))..((((((...(((....................))).)))))).......... (-17.23 = -17.15 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:55 2006