
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,138,851 – 4,138,983 |
| Length | 132 |
| Max. P | 0.994442 |

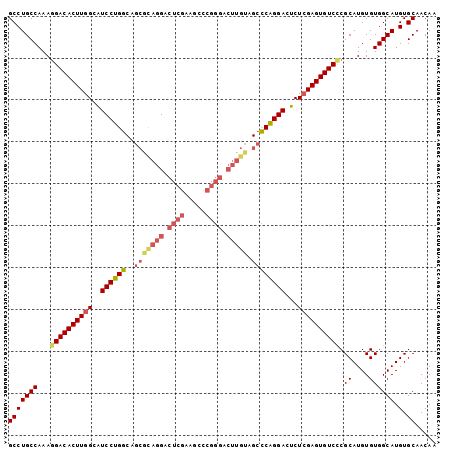
| Location | 4,138,851 – 4,138,953 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -35.49 |
| Energy contribution | -37.05 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4138851 102 + 22407834 GCCUGCCAAAGGACACUUGGCAUCCUGGCAGCGCAGGACUCGAUGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA (((((((...((((((((((..((((((..(((((((.((((.....)))).))))).))))))))...)))))))))).((....)))))).).))..... ( -43.60) >DroSec_CAF1 268286 102 + 1 GCCUGCCAAAUGACACUUGGCAUCCUGGCAGCGCAGGACUCGAAGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA ((((((((.(((((((((((..((((((..(((((((.((((.....)))).))))).))))))))...)))))))))....))...))))).).))..... ( -40.20) >DroSim_CAF1 271926 102 + 1 GCCUGCCAAAGGACACUUGGCAUCCUGGCAGCGCAGGACUCGAAGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA (((((((...((((((((((..((((((..(((((((.((((.....)))).))))).))))))))...)))))))))).((....)))))).).))..... ( -43.60) >DroEre_CAF1 282282 102 + 1 GCCUGCCAAAGGACACUUGGCGUCCUGGCAGCGUAGGACUCGAAGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA (((((((...((((((((((.((((((((.(((.((..((((.....)))).))))).).))))))).).))))))))).((....)))))).).))..... ( -43.80) >DroYak_CAF1 286869 102 + 1 GCCUGCCAAAGGACACUUGGCGUCCUGGCAGCACAGGACUCGCAGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA (((((((...((((((((((.(((((((..(((((((.((((.....)))).))))).))))))))).).))))))))).((....)))))).).))..... ( -47.10) >DroAna_CAF1 315863 87 + 1 GCCUGCCAAAGGACACUUAGCAUCCUGGCAGGCCAGGACUCU---------------UGGUCGGGACUCUCGAGUGUCUCGCAUGUGUGGCAUGUGCAACAA ((((((((..(((.........)))))))))))...(((...---------------..)))(((((........)))))(((((((...)))))))..... ( -32.10) >consensus GCCUGCCAAAGGACACUUGGCAUCCUGGCAGCGCAGGACUCGAAGCCCGGGACUUGUAGCCCAGGACUCUCGAGUGUCCCGCAUGUGUGGCAUGUGCAACAA (((((((...((((((((((..((((((..(((((((.((((.....)))).))))).))))))))...)))))))))).((....)))))).).))..... (-35.49 = -37.05 + 1.56)


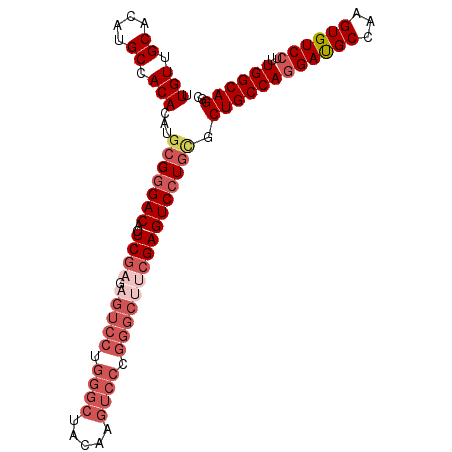
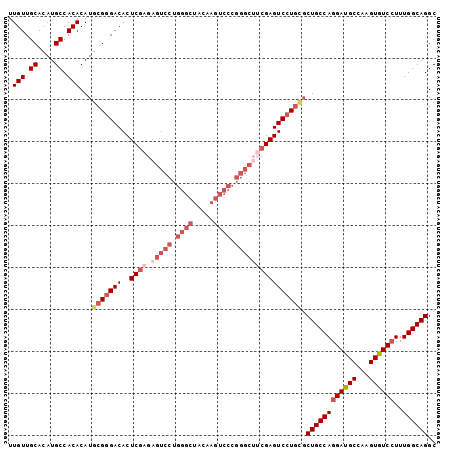
| Location | 4,138,851 – 4,138,953 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -43.55 |
| Consensus MFE | -37.13 |
| Energy contribution | -39.63 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4138851 102 - 22407834 UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCAUCGAGUCCUGCGCUGCCAGGAUGCCAAGUGUCCUUUGGCAGGC .(((.((....)).)))...(((((((..((((..((((.((((.....)))).)))).))))))))))).((((((((((((...))))))..)))))).. ( -46.70) >DroSec_CAF1 268286 102 - 1 UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCUGCCAGGAUGCCAAGUGUCAUUUGGCAGGC .(((.((....)).)))...(((((((..((((.(((((.((((.....)))).)))))))))))))))).((((((.(((((...)))))...)))))).. ( -45.40) >DroSim_CAF1 271926 102 - 1 UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCUGCCAGGAUGCCAAGUGUCCUUUGGCAGGC .(((.((....)).)))...(((((((..((((.(((((.((((.....)))).)))))))))))))))).((((((((((((...))))))..)))))).. ( -47.70) >DroEre_CAF1 282282 102 - 1 UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUACGCUGCCAGGACGCCAAGUGUCCUUUGGCAGGC ..(((.(.((((.....)))).).)))((((((.(((((.((((.....)))).)))))))))))......((((((((((((...))))))..)))))).. ( -45.60) >DroYak_CAF1 286869 102 - 1 UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUGCGAGUCCUGUGCUGCCAGGACGCCAAGUGUCCUUUGGCAGGC .(((.((....)).)))...(((((((..(((..(((((.((((.....)))).))))).)))))))))).((((((((((((...))))))..)))))).. ( -46.90) >DroAna_CAF1 315863 87 - 1 UUGUUGCACAUGCCACACAUGCGAGACACUCGAGAGUCCCGACCA---------------AGAGUCCUGGCCUGCCAGGAUGCUAAGUGUCCUUUGGCAGGC ..((..(.((((.....)))).)..))((((..(.((....))).---------------.))))....((((((((((((((...))))))..)))))))) ( -29.00) >consensus UUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGCUGCCAGGAUGCCAAGUGUCCUUUGGCAGGC .(((.((....)).)))...(((((((..((((.(((((.((((.....)))).)))))))))))))))).((((((((((((...))))))..)))))).. (-37.13 = -39.63 + 2.50)

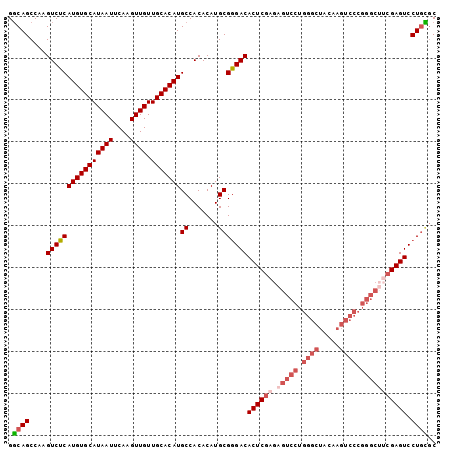
| Location | 4,138,881 – 4,138,983 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -34.81 |
| Energy contribution | -36.62 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4138881 102 - 22407834 GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCAUCGAGUCCUGCGC .((......((..((((((((((((....)))).))))))))..))....(((((((..((((..((((.((((.....)))).)))).))))))))))))) ( -42.90) >DroSec_CAF1 268316 102 - 1 GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGC .((......((..((((((((((((....)))).))))))))..))....(((((((..((((.(((((.((((.....)))).)))))))))))))))))) ( -43.90) >DroSim_CAF1 271956 102 - 1 GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGC .((......((..((((((((((((....)))).))))))))..))....(((((((..((((.(((((.((((.....)))).)))))))))))))))))) ( -43.90) >DroEre_CAF1 282312 102 - 1 GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUACGC .(((.....((..((((((((((((....)))).))))))))..))...)))(((((..((((.(((((.((((.....)))).)))))))))))))).... ( -39.50) >DroYak_CAF1 286899 102 - 1 GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUGCGAGUCCUGUGC .((......((..((((((((((((....)))).))))))))..))....(((((((..(((..(((((.((((.....)))).))))).)))))))))))) ( -40.30) >DroAna_CAF1 315893 87 - 1 GCCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGAGACACUCGAGAGUCCCGACCA---------------AGAGUCCUGGCC (((((....((((((((((((((((....)))).)))))))((.......)))))))((((..(.((....))).---------------.)))).))))). ( -29.20) >consensus GGCAGCCAAGUCUCAUGUGCAUAAUUCAAGUUGUUGCACAUGCCACACAUGCGGGACACUCGAGAGUCCUGGGCUACAAGUCCCGGGCUUCGAGUCCUGCGC .((((....((((((((((((((((....)))).)))))))((.......)))))))((((((.(((((.((((.....)))).))))))))))).)))).. (-34.81 = -36.62 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:52 2006