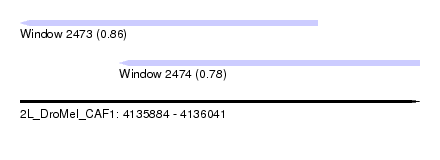
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,135,884 – 4,136,041 |
| Length | 157 |
| Max. P | 0.864452 |

| Location | 4,135,884 – 4,136,001 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 97.95 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -29.02 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
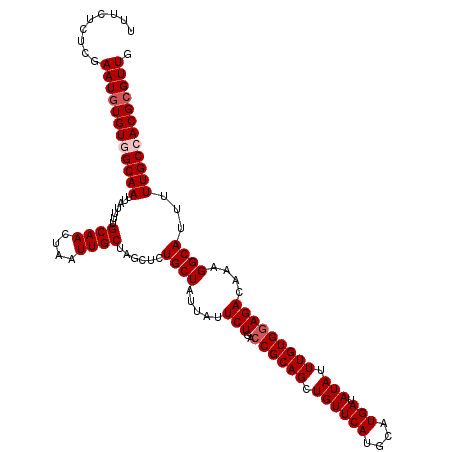
>2L_DroMel_CAF1 4135884 117 - 22407834 UAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGGCAAAUUGG ......(((((...((((..((((((.((((((....))).))).)))))).......((((....))))..(((((((((.((...)).)))))))))))))...)))))...... ( -32.00) >DroSec_CAF1 265347 117 - 1 UAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGGCAAAUUGG ......(((((...((((..((((((.((((((....))).))).)))))).......((((....))))..(((((((((.((...)).)))))))))))))...)))))...... ( -32.00) >DroSim_CAF1 268987 117 - 1 UAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGGCAAAUUGG ......(((((...((((..((((((.((((((....))).))).)))))).......((((....))))..(((((((((.((...)).)))))))))))))...)))))...... ( -32.00) >DroEre_CAF1 279289 117 - 1 UGGCGGUGCUAUUACUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGAAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGUCAAAUUGG ..(((((.(.........)))))).((.......))................((((((((((....))))..(((((((((.((...)).))))))))).....))))))....... ( -31.10) >DroYak_CAF1 283891 117 - 1 UAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGGCAAAUUGG ......(((((...((((..((((((.((((((....))).))).)))))).......((((....))))..(((((((((.((...)).)))))))))))))...)))))...... ( -32.00) >consensus UAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUGAUUUUGGCUCAUAAUUAACGCAGAACUUUGGCAAAUUGG ......(((((...((((..((((((.((((((....))).))).)))))).......((((....))))..(((((((((.((...)).)))))))))))))...)))))...... (-29.02 = -29.62 + 0.60)


| Location | 4,135,923 – 4,136,041 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4135923 118 - 22407834 UUUCUCUCGAAUGUGUGGCAAUUAUUUUGCAACUAAUUGCUAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUG .........((((((((((((.......((((....))))......((((.....(((..((((((.((((((....))).))).)))))))))....))))...)))))))))))). ( -31.60) >DroSec_CAF1 265386 118 - 1 UUGCUCUCGAAUGUGUGGCAAUUAUUUUGCAACUAAUUGCUAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUG .........((((((((((((.......((((....))))......((((.....(((..((((((.((((((....))).))).)))))))))....))))...)))))))))))). ( -31.60) >DroSim_CAF1 269026 118 - 1 UUGCUCUCGAAUGUGUGGCAAUUAUUUUGCAACUAAUUGCUAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUG .........((((((((((((.......((((....))))......((((.....(((..((((((.((((((....))).))).)))))))))....))))...)))))))))))). ( -31.60) >DroEre_CAF1 279328 118 - 1 UUUCCCUCGAAUAUGUAGCAAUUAUUUUGCAACUAAUUGCUGGCGGUGCUAUUACUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGAAGACAAAGGCAUUUUUGCCACGCGUUG ........(((((.((.((((.....)))).)).....((((.((((.(.........))))))))))))))((((........(((((....)))))((((....))))..)))).. ( -29.50) >DroYak_CAF1 283930 118 - 1 UUUCCCUCGAAUGUGUAGCAAUUAUUUUGCAACUAAUUGCUAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUG (((((..(((((((((.((((.....)))).......(((.(((.((((.............))))..)))...)))..))))))))).)))))....((((....))))........ ( -28.82) >consensus UUUCUCUCGAAUGUGUGGCAAUUAUUUUGCAACUAAUUGCUAGCUCUGCUAUUAUUCUGACCGCAGCUGUUCAUGCAUGAUAUAUUUGUGGAGACAAAGGCAUUUUUGCCACGCGUUG .........((((((((((((.......((((....))))......((((.....(((..((((((.((((((....))).))).)))))))))....))))...)))))))))))). (-27.12 = -27.92 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:47 2006