
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
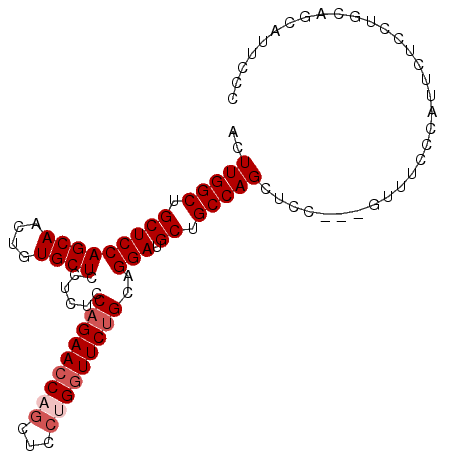
| Location | 4,128,549 – 4,128,643 |
| Length | 94 |
| Max. P | 0.999928 |

| Location | 4,128,549 – 4,128,643 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -23.42 |
| Energy contribution | -24.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
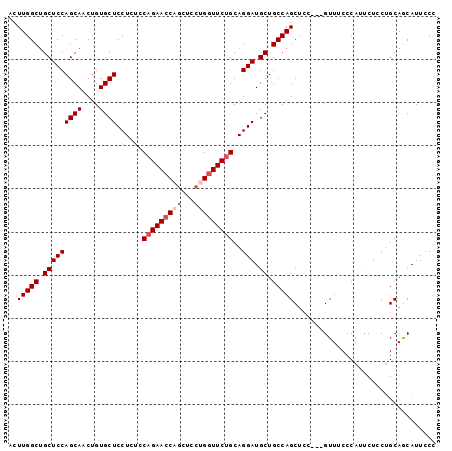
>2L_DroMel_CAF1 4128549 94 + 22407834 --UUGGCUGCUCCAGCAACUGUGCUCCUCUUCAGAACCAGCUCCUGAUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCUGCAACAUUCCC --.((((.(((((((((....))))......(((((.(((...))).)))))..))).)).))))((...---...............))......... ( -21.37) >DroSec_CAF1 257515 96 + 1 ACUUGGCUGCUCCAGCAACUGUGCUCCUCUCCAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCUGCAGCAUUCCC ...((((.(((((((((....))))......(((((((((...)))))))))..))).)).))))(((.(---(.............)).)))...... ( -29.22) >DroSim_CAF1 261188 96 + 1 ACUUGGCUGCUCCAGCAACUGUGCUCCUCUCCAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAUUCUCCGGCAGCAUUCCC ...((((.(((((((((....))))......(((((((((...)))))))))..))).)).))))(((((---(............))).)))...... ( -34.00) >DroEre_CAF1 268669 94 + 1 ACUUGGCUGCUCCAGCAACUGUGCUCCU--CCAGAACCUGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC---GUUUCCCAACCUCCUGCAGCAUUCCC .....(((((..(((((.......((((--.(((((((.......))))))).)))))))))........---(((....))).....)))))...... ( -28.61) >DroYak_CAF1 276113 97 + 1 ACUUGGCCGCUCCAGCAACUGUGCUCCU--CCAGAACCUCCUCCUGGUUCAGCAGGAUGCUGCCAGCUUCCCAGUUACCCAACCUCCUGCAACAUUCCC ...((((.(((((((((....))))...--...(((((.......)))))....))).)).))))((......(((....))).....))......... ( -23.30) >consensus ACUUGGCUGCUCCAGCAACUGUGCUCCUCUCCAGAACCAGCUCCUGGUUCUGCAGGAUGCUGCCAGCUCC___GUUUCCCAUUCUCCUGCAGCAUUCCC ..(((((.(((((((((....))))......(((((((((...)))))))))..))).)).)))))................................. (-23.42 = -24.42 + 1.00)

| Location | 4,128,549 – 4,128,643 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -33.84 |
| Energy contribution | -34.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
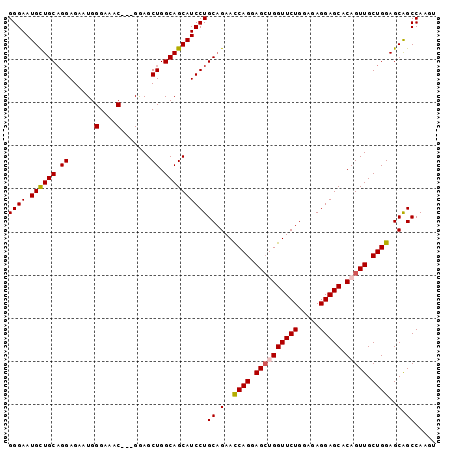
>2L_DroMel_CAF1 4128549 94 - 22407834 GGGAAUGUUGCAGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAAUCAGGAGCUGGUUCUGAAGAGGAGCACAGUUGCUGGAGCAGCCAA-- ((...((((((((((.....(....)---...((.....)).))))))....((((.((((((((((.....))))).))))).)))))))).))..-- ( -34.70) >DroSec_CAF1 257515 96 - 1 GGGAAUGCUGCAGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUGGAGAGGAGCACAGUUGCUGGAGCAGCCAAGU ((...((((((((((.....(....)---...((.....)).))))))....((((.((((((((((.....))))).))))).)))))))).)).... ( -39.90) >DroSim_CAF1 261188 96 - 1 GGGAAUGCUGCCGGAGAAUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUGGAGAGGAGCACAGUUGCUGGAGCAGCCAAGU ((((.(((((((((....(.(....)---.)..)))))))))))))((.(..((((.((((((((((.....))))).))))).))))..).))..... ( -44.50) >DroEre_CAF1 268669 94 - 1 GGGAAUGCUGCAGGAGGUUGGGAAAC---GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCAGGUUCUGG--AGGAGCACAGUUGCUGGAGCAGCCAAGU .....((((((...((.((.(....)---.)).)).))))))((((.(((((((.......))))))).--)))).....(((((....)))))..... ( -40.10) >DroYak_CAF1 276113 97 - 1 GGGAAUGUUGCAGGAGGUUGGGUAACUGGGAAGCUGGCAGCAUCCUGCUGAACCAGGAGGAGGUUCUGG--AGGAGCACAGUUGCUGGAGCGGCCAAGU ............((.(.((.(((((((((((.((.....)).)))((((...((((((.....))))))--...)))))))))))).)).)..)).... ( -32.80) >consensus GGGAAUGCUGCAGGAGAAUGGGAAAC___GGAGCUGGCAGCAUCCUGCAGAACCAGGAGCUGGUUCUGGAGAGGAGCACAGUUGCUGGAGCAGCCAAGU ((((.((((((.((......(....).......)).))))))))))((.(..((((.((((((((((.....))))).))))).))))..).))..... (-33.84 = -34.04 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:40 2006