
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,118,973 – 4,119,080 |
| Length | 107 |
| Max. P | 0.907772 |

| Location | 4,118,973 – 4,119,080 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -16.81 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4118973 107 + 22407834 CAAAACCACUGGCAACAGUGGCUACGCCGCAAAUAAAAGAAUAAUAA---UAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCAGUGAAACAGCGGCAG---UCAGCG ......(((((....)))))((((((((((.................---.........((..........))....(....)..............))))).)---).))). ( -24.80) >DroGri_CAF1 352229 89 + 1 AA-AACCAG---------------CGGCGCUAAUAAAUG--UUAUAA---UCAUAAUGAGCAAAUGUUUAAACAGUAGCAAAGGGAGCAGUGCAAGGGCGGAAG---GGGUGG ..-.(((..---------------(.((.((..((((((--((....---((.....))...)))))))).......(((..(....)..))).)).)).)...---.))).. ( -16.30) >DroSec_CAF1 247951 107 + 1 CAAAACCACUGGCAACAGUGGCUGCGCCGCAAAUAAAAGAAUAAUAA---UAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCAGUGAAGCAGCGGCAG---UCAGGG .....((((((....))))))(((((((((.................---.........((..........)).((.....(((....)))...)).))))).)---.))).. ( -27.20) >DroSim_CAF1 251497 106 + 1 CAAAACCACUGGCAACAGUGGCUGCGCCGCAAAUAAAAGAAUAAUAA---UAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCAGCGAAGCAGCGGCAG----CAGCG ......(((((....)))))((((((((((.................---.........((..........)).((.((.......)).))......))))).)----)))). ( -32.20) >DroEre_CAF1 256925 110 + 1 GAAAACCACUGGCAACAGUGGCUACGCCGCAAAUAAAAGAAUAAUAAUAAUAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCGGUGAAACAGUGGCAG---UCAGGG .....((((((....))))))....(((((.............................((..........)).(((((.......)))))......)))))..---...... ( -27.40) >DroYak_CAF1 262359 110 + 1 CAAAACCACUGGCAACAGUGGCCAUGCCGCAAAUAAAAGAAUAAUAA---UAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCUGUGAAACGGCGGCGGCGAGCAGAG .....((((((....))))))...((((((.................---.........((..........)).((((...((......))....)))).))))))....... ( -32.50) >consensus CAAAACCACUGGCAACAGUGGCUACGCCGCAAAUAAAAGAAUAAUAA___UAAUAAUGAGCAAAUAUUUAAGCAGCCGGGAACUUAUCAGUGAAACAGCGGCAG___UCAGGG .....((((((....))))))....(((((.............................((..........)).((.((.......)).))......)))))........... (-16.81 = -17.37 + 0.56)

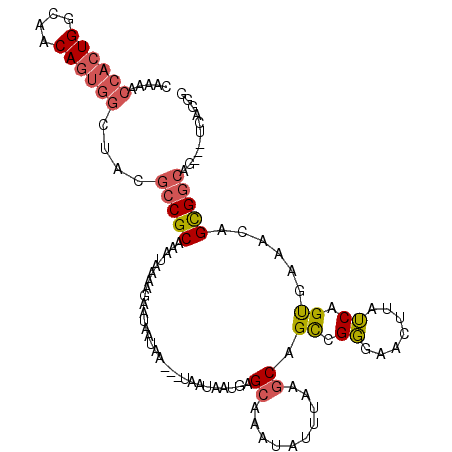
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:34 2006