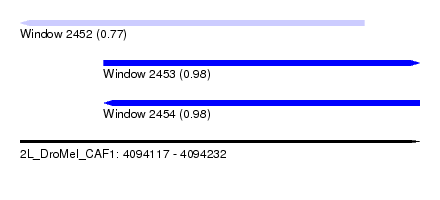
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,094,117 – 4,094,232 |
| Length | 115 |
| Max. P | 0.982538 |

| Location | 4,094,117 – 4,094,216 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
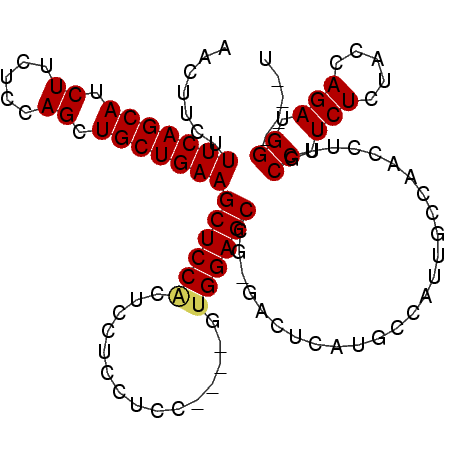
>2L_DroMel_CAF1 4094117 99 - 22407834 AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCGCC----AUGGAGCCGAGACUCAUGCCAUUGCUAACCUUGCCUUCUCUACCAGAUGG----U ......((((((((.((.....)).))))))))(((..((((......----..))))..)))......((((((((.......))...........)))))----) ( -21.90) >DroSec_CAF1 223463 98 - 1 AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC----UUGGAGCGG-GACUCAUGCCAUUGCGAACCUUGCCUUCUCUACCAGAUGG----U .......(((((((.((.....)).)))))))((((((..........----.))))))((-..(.((......)).)..))..(((.(((.....))).))----) ( -24.30) >DroSim_CAF1 226347 98 - 1 AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC----UUGGAGCCG-GACUCAUGCCAUUGCCAACCUUGCCUUCUCUACCAGAUGG----U .......(((((((.((.....)).)))))))((((((..........----.)))))).(-(.(....)))............(((.(((.....))).))----) ( -21.60) >DroEre_CAF1 231972 98 - 1 AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCGCUCCUCCUCC----GUGGAGCCG-GACUCCUGCCAUGCCCAACCUAGCCAUCUCUGCCAGAUGG----U .......(((((((.((.....)).)))))))(((((((.........----))))))).(-(.(....)))............(((((((.....))))))----) ( -29.20) >DroYak_CAF1 237119 106 - 1 AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUUCUUCGGAGGAGCCG-GUCUCCUGCCACUCGCAACCUAGCCAUCUCUACCAGAUGGUUGGU .......(((((((.((.....)).)))))))((....(((((((......)))))))..(-((.....)))....))...((((((((((.....)))))))))). ( -38.50) >consensus AACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC____GUGGAGCCG_GACUCAUGCCAUUGCCAACCUUGCCUUCUCUACCAGAUGG____U .......(((((((.((.....)).)))))))((((((...............))))))..........................((.(((.....))).))..... (-17.82 = -17.90 + 0.08)

| Location | 4,094,141 – 4,094,232 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
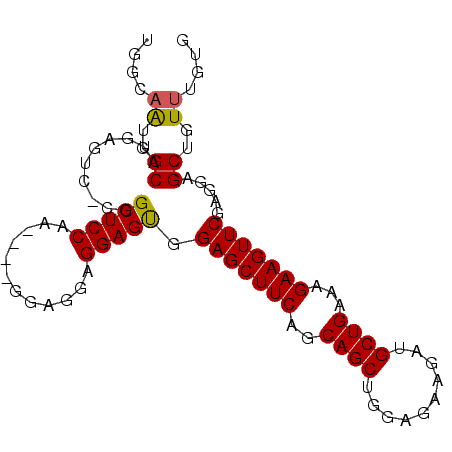
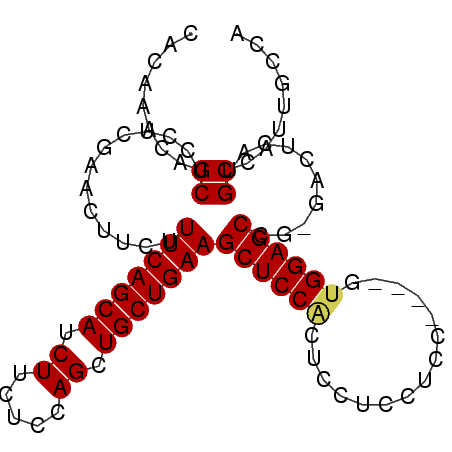
>2L_DroMel_CAF1 4094141 91 + 22407834 UAGCAAUGGCAUGAGUCUCGGCUCCAU----GGCGGAGGAGUGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG ..((((..(((.(..((((((((((((----..(....).)))))))(((((((.((.....)).))))))).......)))))..)))))))). ( -31.70) >DroSec_CAF1 223487 90 + 1 UCGCAAUGGCAUGAGUC-CCGCUCCAA----GGAGGAGGAGUGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG .(((((((((....(..-(((((((..----......)))))))..)(((((((.((.....)).))))))).............)))).))))) ( -31.60) >DroSim_CAF1 226371 90 + 1 UGGCAAUGGCAUGAGUC-CGGCUCCAA----GGAGGAGGAGUGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG ..((((((((...(.((-(..(((...----.)))..))).).(((((((..((((..........))))...))))))).....)))).)))). ( -29.20) >DroEre_CAF1 231996 90 + 1 UGGGCAUGGCAGGAGUC-CGGCUCCAC----GGAGGAGGAGCGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG ...(((.(((((...((-(.(((((..----......))))).(((((((..((((..........))))...)))))))..))).)))))))). ( -32.10) >DroYak_CAF1 237147 94 + 1 UGCGAGUGGCAGGAGAC-CGGCUCCUCCGAAGAAGGAGGAGUGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG .(((((..((.(....)-..((((((((......)))))))).(((((((..((((..........))))...))))))).....))..))))). ( -40.40) >consensus UGGCAAUGGCAUGAGUC_CGGCUCCAA____GGAGGAGGAGUGGAGCUUCAGCAGCUGGAGAAGAUGCUGAAAGAAGUUCGAGGAGCUGUUUGUG ....((..((..........(((((............))))).(((((((..((((..........))))...))))))).....))..)).... (-25.04 = -24.92 + -0.12)

| Location | 4,094,141 – 4,094,232 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4094141 91 - 22407834 CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCGCC----AUGGAGCCGAGACUCAUGCCAUUGCUA ............((((.......(((((((.((.....)).)))))))((((((..........----.))))))))))................ ( -21.90) >DroSec_CAF1 223487 90 - 1 CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC----UUGGAGCGG-GACUCAUGCCAUUGCGA .......((..((..........(((((((.((.....)).)))))))((((((..........----.))))))))-..))..(((....))). ( -22.10) >DroSim_CAF1 226371 90 - 1 CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC----UUGGAGCCG-GACUCAUGCCAUUGCCA .......((.(((..........(((((((.((.....)).)))))))((((((..........----.)))))).)-))))............. ( -19.90) >DroEre_CAF1 231996 90 - 1 CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCGCUCCUCCUCC----GUGGAGCCG-GACUCCUGCCAUGCCCA ......(((.(((..........(((((((.((.....)).)))))))(((((((.........----))))))).)-))...)))......... ( -24.40) >DroYak_CAF1 237147 94 - 1 CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUUCUUCGGAGGAGCCG-GUCUCCUGCCACUCGCA .......................(((((((.((.....)).)))))))((....(((((((......)))))))..(-((.....)))....)). ( -25.90) >consensus CACAAACAGCUCCUCGAACUUCUUUCAGCAUCUUCUCCAGCUGCUGAAGCUCCACUCCUCCUCC____GUGGAGCCG_GACUCAUGCCAUUGCCA ........((.............(((((((.((.....)).)))))))((((((...............))))))..........))........ (-17.30 = -17.38 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:27 2006