| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,090,074 – 4,090,170 |
| Length | 96 |
| Max. P | 0.897617 |

| Location | 4,090,074 – 4,090,170 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -15.07 |
| Energy contribution | -17.10 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4090074 96 + 22407834 UGUCUAAGUGUGUGUGCUCUUAUAUGC--------------UCUG----AAU----AUAUAUUUUGAAUAUCUACACCGAACA--CAAAGUCCGAUAACAAUAGGCACACUCACUUCUCA .....(((((.(((((((..(((((..--------------....----.))----)))..................((.((.--....)).)).........))))))).))))).... ( -18.20) >DroSec_CAF1 219490 120 + 1 UGUCUAAGUGUGAGUGCUCUAGUAGGCGAAUAAUUAUUUUCUCUAUGCUAAUACUCAUACUUUUUGAAUAUCUUCACCGAACACUCAAGGUCCGAUAACAAUGGGCACACUCACUUCUCA ((((((((((((((((.....(((((.(((........)))))))).....)))))))))).......((((...(((..........)))..))))....))))))............. ( -25.80) >DroSim_CAF1 222376 120 + 1 UGUCCAAGUGUGUGCGCUCUAGUAGGCGAAUAAUUAUUUUCUCUAUGCUAAUACUCAUACUUUUUGAAUAUCUUCACCGAACACACAAGGUCCGAUAACAAUGGGCACACUCACUUCUCA .....(((((.(((.(((((((((((.(((........))).)).)))))..................((((...(((..........)))..)))).....)))).))).))))).... ( -24.40) >consensus UGUCUAAGUGUGUGUGCUCUAGUAGGCGAAUAAUUAUUUUCUCUAUGCUAAUACUCAUACUUUUUGAAUAUCUUCACCGAACAC_CAAGGUCCGAUAACAAUGGGCACACUCACUUCUCA .....(((((.((((((((..(((((.(((........))))))))......................((((...(((..........)))..)))).....)))))))).))))).... (-15.07 = -17.10 + 2.03)

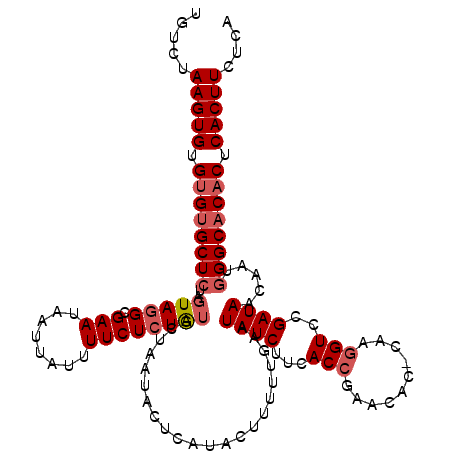
| Location | 4,090,074 – 4,090,170 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.81 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4090074 96 - 22407834 UGAGAAGUGAGUGUGCCUAUUGUUAUCGGACUUUG--UGUUCGGUGUAGAUAUUCAAAAUAUAU----AUU----CAGA--------------GCAUAUAAGAGCACACACACUUAGACA ....(((((.((((((((...(((....)))..((--(((((...(((.((((.....)))).)----)).----..))--------------)))))..)).)))))).)))))..... ( -25.30) >DroSec_CAF1 219490 120 - 1 UGAGAAGUGAGUGUGCCCAUUGUUAUCGGACCUUGAGUGUUCGGUGAAGAUAUUCAAAAAGUAUGAGUAUUAGCAUAGAGAAAAUAAUUAUUCGCCUACUAGAGCACUCACACUUAGACA ((((..((((((((((......((((((((((....).))))))))).((((((((.......)))))))).)).(((.(((........)))..))).....)))))))).)))).... ( -30.30) >DroSim_CAF1 222376 120 - 1 UGAGAAGUGAGUGUGCCCAUUGUUAUCGGACCUUGUGUGUUCGGUGAAGAUAUUCAAAAAGUAUGAGUAUUAGCAUAGAGAAAAUAAUUAUUCGCCUACUAGAGCGCACACACUUGGACA ....(((((.((((((.(....((((((((((....).))))))))).((((((((.......))))))))....(((.(((........)))..)))...).)))))).)))))..... ( -30.90) >consensus UGAGAAGUGAGUGUGCCCAUUGUUAUCGGACCUUG_GUGUUCGGUGAAGAUAUUCAAAAAGUAUGAGUAUUAGCAUAGAGAAAAUAAUUAUUCGCCUACUAGAGCACACACACUUAGACA ....(((((.((((((.((((.(((((((((.......))))))))).)))........((((((((((...................))))))..)))).).)))))).)))))..... (-22.80 = -22.81 + 0.01)

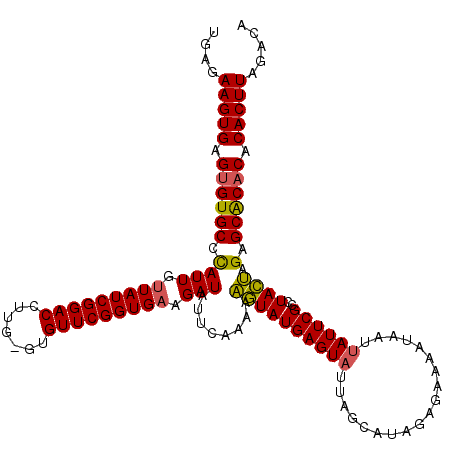
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:24 2006