
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,086,898 – 4,087,054 |
| Length | 156 |
| Max. P | 0.998233 |

| Location | 4,086,898 – 4,086,995 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
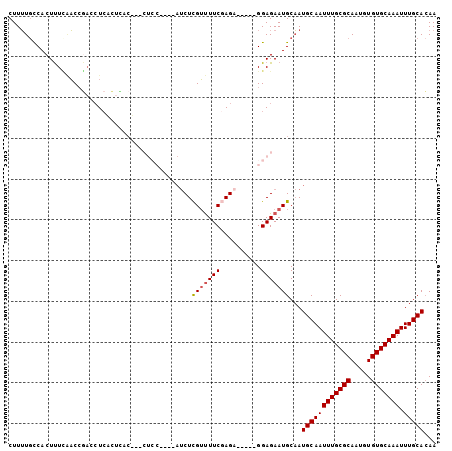
>2L_DroMel_CAF1 4086898 97 + 22407834 CUUUUGCCACUUUCAACAGACCUCACUUCCUCCCUCC---CAUCUCGUUUUCGAGA-----GGAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAA ...((((.....((....)).............((((---..(((((....)))))-----))))...))))((((((((((((.....))))))).)))))... ( -26.00) >DroSec_CAF1 216355 100 + 1 CUUUUGCCACUUUCAACCGUCCUCACUCACUUCCUCCUCAAAUCUCGUUUUCGAGA-----GGAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAA ...((((...........((....))..........(((...(((((....)))))-----.)))...))))((((((((((((.....))))))).)))))... ( -24.50) >DroSim_CAF1 219239 89 + 1 CUUUUGCCACUUUUAACCGACCUCACUCAC-----------AUCUCGUUUUCCAGA-----GGAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAA ...((((...........((......))..-----------.((((.((.....))-----.))))..))))((((((((((((.....))))))).)))))... ( -20.40) >DroEre_CAF1 224735 96 + 1 CUUUUCCCACUUUCCACCAACCUCUUUUCCA-UCUCC---CAUCUCGUUUUCGAGC-----GGAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAA ...............................-(((((---...((((....)))).-----)))))......((((((((((((.....))))))).)))))... ( -24.40) >DroAna_CAF1 260610 86 + 1 CUUGUGUCACUCUAGCCCUAUAU-------------------CCUUGGCUUCCAGAGAGCUGGAGAAUGCCAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCAGAA ......((...............-------------------...((((((((((....))))))...))))((((((((((((.....))))))).))))))). ( -25.60) >consensus CUUUUGCCACUUUCAACCGACCUCACUCAC___CUCC____AUCUCGUUUUCGAGA_____GGAGAAUGCAAUGCAAUUUGCGCAAUGUGUGCAAAUUUGCACAA .............................................(((((((..........)))))))...((((((((((((.....))))))).)))))... (-16.32 = -16.56 + 0.24)


| Location | 4,086,959 – 4,087,054 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.60 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -8.77 |
| Energy contribution | -8.46 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
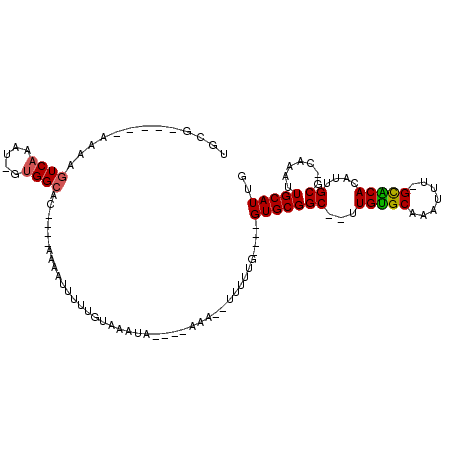
>2L_DroMel_CAF1 4086959 95 - 22407834 UGCGUUGGCAAAAGUCAAAU-UUGGCAC----GAAAAUUUUGUAAAUA----AAA--UUUUUG----GUGCGGC--UUGUGCAAAUUU-GCACACAUUGCG-CAAAUUGCAUUG .......((((..((((...-.)))).(----((((((((((....))----)))--))))))----((((((.--.((((((....)-)))))..)))))-)...)))).... ( -28.40) >DroVir_CAF1 319904 89 - 1 UGU-------CGAGUCAAAU-GUGGCAG----CAAAUUUCUGUAAAUA----AGGA-AUUUUU----GUGCGGC--UUGCGCAAAUUU-GUGCAGAUUGCA-CAAAUUGCAUUG ...-------...((((...-.)))).(----(((((((((.......----))))-)).(((----((((((.--(((((((....)-)))))).)))))-)))))))).... ( -28.20) >DroPse_CAF1 340021 108 - 1 UGGGGGCGAAAAAGUCAAAUUUUGGCACUCCAAAAAUUUUUGUAAAUACGAAAAA--UGUGUGCAAUGUGCGGC--UUGUGCAAAUUU-GCACACAUUGCG-CAAAUUGCAUUG (((((((.((((.......)))).)).)))))....(((((((....))))))).--...(((((((((((((.--.((((((....)-)))))..)))))-))..)))))).. ( -35.00) >DroGri_CAF1 313067 96 - 1 UGCG----CCCGACUCUAAU-GUGGCAG----CAAAGUUUUGUAAAUA----AAAAUUUGUGU----GUGCGGCUCUUGUGCCAAUUUGGUGCAGUUUGCG-CAGAUUGCAUUG ((((----(..(((......-((.((((----(((((((((.......----))))))).)))----.))).))...((..((.....))..))))).)))-)).......... ( -25.70) >DroMoj_CAF1 363056 91 - 1 UGC-------GAAGUCAAAU-GUGGCUG----AAAAUUUUUGUAAAUA----AGGAAAUUUUU----GUGCGGC--UUGUGCAAAUUU-GUGCAGAUUGCGCCAAAUUGCAUUG (((-------(((((((...-.)))))(----(((((((((.......----.))))))))))----((((((.--(((..((....)-)..))).))))))....)))))... ( -24.30) >DroPer_CAF1 340818 108 - 1 UGGGGGCGAAAAAGUCAAAUUUUGGCACUCCAAAAAUUUUUGUAAAUACGAAAAA--UGUGUGCAAUGUGCGGC--UUGUGCAAAUUU-GCACACAUUGCG-CAAAUUGCAUUG (((((((.((((.......)))).)).)))))....(((((((....))))))).--...(((((((((((((.--.((((((....)-)))))..)))))-))..)))))).. ( -35.00) >consensus UGCG_____AAAAGUCAAAU_GUGGCAC____AAAAUUUUUGUAAAUA____AAA__UUUUUG____GUGCGGC__UUGUGCAAAUUU_GCACACAUUGCG_CAAAUUGCAUUG .............((((.....)))).........................................(((((((...(((((.......)))))....)).......))))).. ( -8.77 = -8.46 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:20 2006