
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,061,106 – 4,061,344 |
| Length | 238 |
| Max. P | 0.999875 |

| Location | 4,061,106 – 4,061,224 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061106 118 + 22407834 CCAUAGAGAUAAUAUUAAUAGCCGACCGCCCAAGUGGUCAGCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCGGCCAGCAUUUGCGUGAAAAG ....................((.((((((....)))))).))(((((.-.(((.....)))......)))))..((((((((.............))))))))((....))........ ( -32.72) >DroSec_CAF1 193377 118 + 1 CCAUAGAGAUAAUAUUAAUAGCCGGCCGCCCAAGUGGUCAGCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAGCUUUUGCGUGAAAAG ....................((.((((((....)))))).))(((((.-.(((.....)))......)))))..((((((((.............))))))))((....))........ ( -33.02) >DroSim_CAF1 194899 118 + 1 CCAUAGAGAUAAUAUUAAUAGCCGACCGCCCAAGUGGUCAGCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAG ....................((.((((((....)))))).))(((((.-.(((.....)))......)))))..((((((((.............))))))))((....))........ ( -33.42) >DroEre_CAF1 201561 118 + 1 CCAUACAGAUAAUAUUAAUAGCCGACCGCCCAAGUGGUCAGCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAACCAGCAUUUGCGUGAAAAG .((..(((((..(((((..((((((((((....)))))).........-.(((.....)))....))))..)))))(((((.....(((.......))).))))))))))..))..... ( -33.00) >DroYak_CAF1 206552 119 + 1 CCAUAGAGAUAAUAUUAAUAGCCGACCGCCCAAGUGGUCAGCAGGGGAAAAUCGAAAAGGUAAGCGACUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAG ....................((.((((((....)))))).))(((((....(((..........))))))))..((((((((.............))))))))((....))........ ( -33.32) >consensus CCAUAGAGAUAAUAUUAAUAGCCGACCGCCCAAGUGGUCAGCAGGGGA_AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAG ....................((.((((((....)))))).))(((((...(((.....)))......)))))..((((((((.............))))))))((....))........ (-32.40 = -32.28 + -0.12)

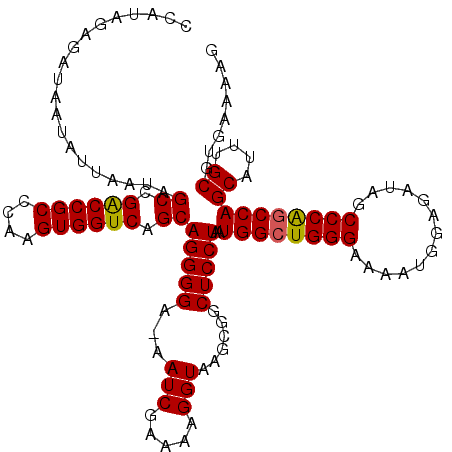

| Location | 4,061,146 – 4,061,264 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -33.57 |
| Energy contribution | -34.65 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061146 118 + 22407834 GCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCGGCCAG-CAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGC ........-..((((((.(((((((((((.....((((((((.............)))))))).-.....((((((.......)))))).....))))))))......))).)))))).. ( -41.02) >DroSec_CAF1 193417 118 + 1 GCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAG-CUUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGC ((((.((.-..((((.((((((............((((((((.............))))))))(-((((.((((((.......))))))...))))))))))).)))).)).)))).... ( -40.52) >DroSim_CAF1 194939 118 + 1 GCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAG-CAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGC ((((.((.-.(((.....)))((((((((.....((((((((.............)))))))).-.....((((((.......)))))).....)))))))).......)).)))).... ( -39.92) >DroEre_CAF1 201601 118 + 1 GCAGGGGA-AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAACCAG-CAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGC ........-..((((((.(((((((((((....((((((((.....(((.......))).))))-)....((((((.......)))))))))..))))))))......))).)))))).. ( -37.50) >DroYak_CAF1 206592 119 + 1 GCAGGGGAAAAUCGAAAAGGUAAGCGACUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAG-CAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGC ...........((((((.(((((((..........(((((((.............)))))))((-(.((.((((((.......))))))...)).)))))))......))).)))))).. ( -35.82) >DroAna_CAF1 238920 102 + 1 ---------AAUCCGAAAGGUAAGCGGGUCUUAAUGGCCC--------CAGAUAGCC-AGCCAGACAUUUGCGUGAAAAGAUGUCACGCCAUAAACUUCCUUUAUUGAACCAUUUCGAGA ---------..((((((((((......((((...((((..--------......)))-)...))))....((((((.......))))))...................))).))))).)) ( -26.00) >consensus GCAGGGGA_AAUCGAAAAGGUAAGCGGCUCCUAAUGGCUGGGAAAAUGGAGAUAGCCCAGCCAG_CAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGC ...........((((((.(((((((((((.....((((((((.............)))))))).......((((((.......)))))).....))))))))......))).)))))).. (-33.57 = -34.65 + 1.09)


| Location | 4,061,146 – 4,061,264 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -29.91 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061146 118 - 22407834 GCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUG-CUGGCCGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGCCGCUUACCUUUUCGAUU-UCCCCUGC ..((((((.(((......((((.(((((...((((((.......)))))).....-.((((.(((..(.......)..))).))))...))))).))))))).))))))..-........ ( -35.20) >DroSec_CAF1 193417 118 - 1 GCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAAG-CUGGCUGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGCCGCUUACCUUUUCGAUU-UCCCCUGC ..(((.((.(((......((((.(((((...((((((.......)))))).....-.((((((((..(.......)..))))))))...))))).))))))).)).)))..-........ ( -35.40) >DroSim_CAF1 194939 118 - 1 GCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUG-CUGGCUGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGCCGCUUACCUUUUCGAUU-UCCCCUGC ..(((.((.(((......((((.(((((...((((((.......)))))).....-.((((((((..(.......)..))))))))...))))).))))))).)).)))..-........ ( -35.40) >DroEre_CAF1 201601 118 - 1 GCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUG-CUGGUUGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGCCGCUUACCUUUUCGAUU-UCCCCUGC ..((((((.(((......((((.(((((...((((((.......)))))).....-.((((((((..(.......)..))))))))...))))).))))))).))))))..-........ ( -37.40) >DroYak_CAF1 206592 119 - 1 GCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUG-CUGGCUGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGUCGCUUACCUUUUCGAUUUUCCCCUGC ..((((((.(((......((((.(((((...((((((.......)))))).....-.((((((((..(.......)..))))))))...))))).))))))).))))))........... ( -38.20) >DroAna_CAF1 238920 102 - 1 UCUCGAAAUGGUUCAAUAAAGGAAGUUUAUGGCGUGACAUCUUUUCACGCAAAUGUCUGGCU-GGCUAUCUG--------GGGCCAUUAAGACCCGCUUACCUUUCGGAUU--------- ((.(((((.(((........((.........((((((.......))))))....((((...(-((((.....--------.)))))...))))))....))))))))))..--------- ( -28.70) >consensus GCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUG_CUGGCUGGGCUAUCUCCAUUUUCCCAGCCAUUAGGAGCCGCUUACCUUUUCGAUU_UCCCCUGC ..(((.((.(((......((((.(((((...((((((.......)))))).......((((((((.............))))))))...))))).))))))).)).)))........... (-29.91 = -30.30 + 0.39)
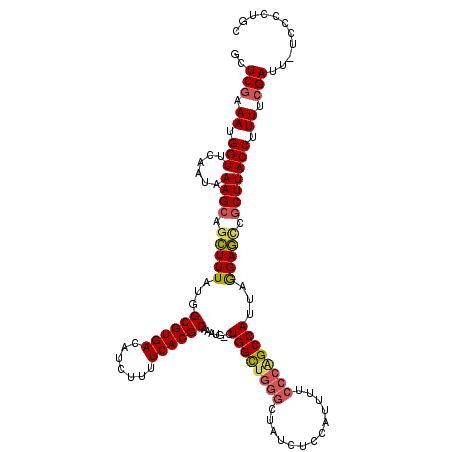

| Location | 4,061,185 – 4,061,304 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -37.10 |
| Energy contribution | -37.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061185 119 + 22407834 GGAAAAUGGAGAUAGCCCGGCCAGCAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCG ((((((((...((((((.(((((((.((.((((((.......))))))...)).))))..........(((....(((((((....))))))))))))).)))).))...)))))))). ( -40.70) >DroSec_CAF1 193456 119 + 1 GGAAAAUGGAGAUAGCCCAGCCAGCUUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCG ((((((((...((((((.((((((((((.((((((.......))))))...)))))))..........(((....(((((((....))))))))))))).)))).))...)))))))). ( -44.80) >DroSim_CAF1 194978 119 + 1 GGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCG ((((((((...((((((.((((.(((...((((((.......)))))).((((((...))))))........)))(((((((....)))))))..)))).)))).))...)))))))). ( -41.20) >DroEre_CAF1 201640 119 + 1 GGAAAAUGGAGAUAGCCCAACCAGCAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAGAUGCGAUUUGGGCGCGGCUCGUAAAUGUUUUCCG (((((.(((.......)))....((((((((((((.......)))).......((((((....(((((....)))))(((((...........))))))))))).))))))))))))). ( -37.00) >DroYak_CAF1 206632 119 + 1 GGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAGAUGCGAUUUGGGCGCGGCUCGUAUAUGUUUUCCG ((((((((...((((((((((.(((.((.((((((.......))))))...)).))))))...(((((....)))))(((((...........)))))).)))).))...)))))))). ( -36.80) >consensus GGAAAAUGGAGAUAGCCCAGCCAGCAUUUGCGUGAAAAGAUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCG ((((((((...((((((....((((.((.((((((.......))))))...)).)))).................(((((((...........))))))))))).))...)))))))). (-37.10 = -37.50 + 0.40)

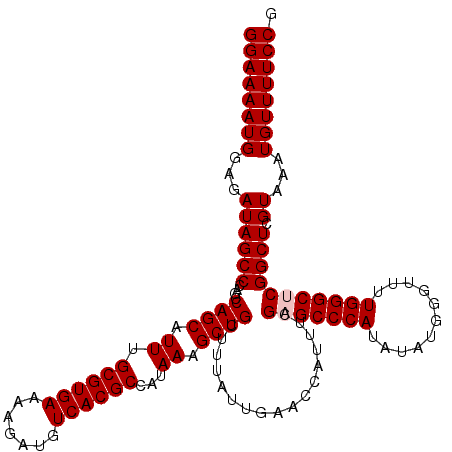
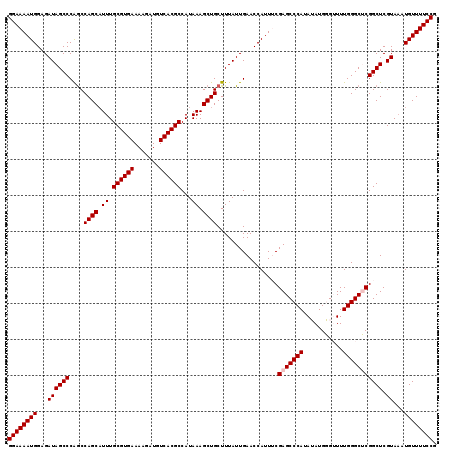
| Location | 4,061,185 – 4,061,304 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -35.02 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061185 119 - 22407834 CGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUGCUGGCCGGGCUAUCUCCAUUUUCC .((((((........(((((((((((...........)))))))....((((.........((((......((((((.......))))))....)))))))))))).......)))))) ( -38.36) >DroSec_CAF1 193456 119 - 1 CGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAAGCUGGCUGGGCUAUCUCCAUUUUCC .((((((........(((((((((((...........)))))))...............(((((((((...((((((.......)))))).)))))).))).)))).......)))))) ( -41.36) >DroSim_CAF1 194978 119 - 1 CGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUGCUGGCUGGGCUAUCUCCAUUUUCC .((((((........(((((((((((...........))))))).................(((((.....((((((.......)))))).......))))))))).......)))))) ( -38.56) >DroEre_CAF1 201640 119 - 1 CGGAAAACAUUUACGAGCCGCGCCCAAAUCGCAUCUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUGCUGGUUGGGCUAUCUCCAUUUUCC .(((((((((((.(((((((((.......))).......)))))))))))(((((((....((((......((((((.......))))))....)))))))))))........)))))) ( -37.91) >DroYak_CAF1 206632 119 - 1 CGGAAAACAUAUACGAGCCGCGCCCAAAUCGCAUCUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUGCUGGCUGGGCUAUCUCCAUUUUCC .((((((........(((((.(((((...........))))).).................(((((.....((((((.......)))))).......))))))))).......)))))) ( -34.06) >consensus CGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAUCUUUUCACGCAAAUGCUGGCUGGGCUAUCUCCAUUUUCC .((((((........(((((((((((...........))))))).................((((......((((((.......))))))....))))....)))).......)))))) (-35.02 = -35.42 + 0.40)

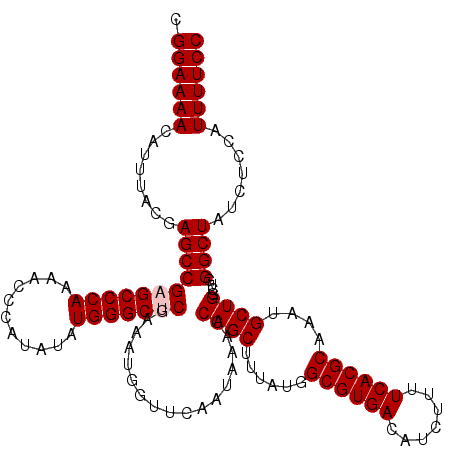
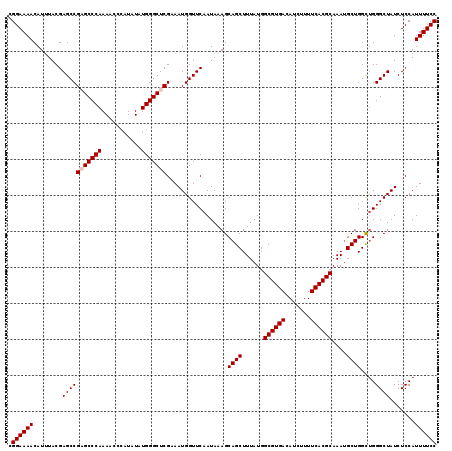
| Location | 4,061,224 – 4,061,344 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -35.69 |
| Consensus MFE | -30.76 |
| Energy contribution | -31.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061224 120 + 22407834 AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCGAGUGCUUUUCCUUGAUAGCAGCGACUUGACAAAUGUCUUG .(((((.........(((((((((..(((......((((((((...........))))))))(((((.((.....)).)))))....)))..))).))))))....)))))......... ( -34.32) >DroSec_CAF1 193495 120 + 1 AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCGAGUGCUUUUCCUUGAUAGCAGCGACUUGACAAAUGCCCAG .(((((.........(((((((((..(((......((((((((...........))))))))(((((.((.....)).)))))....)))..))).))))))....)))))......... ( -34.42) >DroSim_CAF1 195017 120 + 1 AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUGCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCGAGUGCUUUUCCUUGAUAGCAGCGACUUGACAAAUGCCCUG .(((((.........(((((((((..(((......((((((((...........))))))))(((((.((.....)).)))))....)))..))).))))))....)))))......... ( -34.42) >DroEre_CAF1 201679 120 + 1 AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAGAUGCGAUUUGGGCGCGGCUCGUAAAUGUUUUCCGAGUGGUUUUCCUUGAUAGCAGCGGCUUGACAAAUGUCCGG .(((((.(((.....(((((((((..(((((....((.(((((...........))))).))(((((.((.....)).))))))))))....))).))))))))).)))))......... ( -40.30) >DroYak_CAF1 206671 120 + 1 AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAGAUGCGAUUUGGGCGCGGCUCGUAUAUGUUUUCCGAGUGCUUUUCCUUGAUAACAGCGGCUUGACAAAUGCCCUG .(((((.(((....((((((....(((((....)))))(((((...........)))))))))))......((((...((((........))))..))))..))).)))))......... ( -35.00) >consensus AUGUCACGCCAUAAAGCUGCUUUAUUGAACCAUUUCGAGCCCAUAUAUGGGUUUUGGGCUCGGCUCGUAAAUGUUUUCCGAGUGCUUUUCCUUGAUAGCAGCGACUUGACAAAUGCCCUG .(((((.........(((((((((..(((......((((((((...........))))))))(((((...........)))))....)))..))).))))))....)))))......... (-30.76 = -31.36 + 0.60)

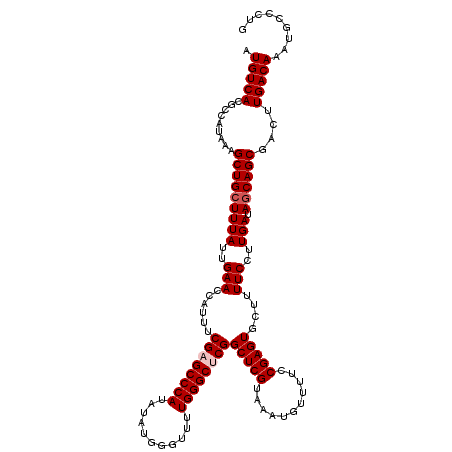
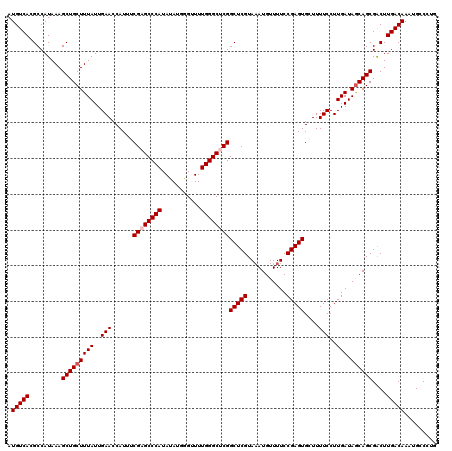
| Location | 4,061,224 – 4,061,344 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -33.82 |
| Energy contribution | -33.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4061224 120 - 22407834 CAAGACAUUUGUCAAGUCGCUGCUAUCAAGGAAAAGCACUCGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU ...(((....)))..(((((.(((((.((((....((.((((...........)))).((((((((...........))))))))...............))..)))))))))))))).. ( -36.90) >DroSec_CAF1 193495 120 - 1 CUGGGCAUUUGUCAAGUCGCUGCUAUCAAGGAAAAGCACUCGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU ...(((....)))..(((((.(((((.((((....((.((((...........)))).((((((((...........))))))))...............))..)))))))))))))).. ( -35.40) >DroSim_CAF1 195017 120 - 1 CAGGGCAUUUGUCAAGUCGCUGCUAUCAAGGAAAAGCACUCGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGCAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU ...(((....)))..(((((.(((((.((((....((.((((...........)))).((((((((...........))))))))...............))..)))))))))))))).. ( -35.40) >DroEre_CAF1 201679 120 - 1 CCGGACAUUUGUCAAGCCGCUGCUAUCAAGGAAAACCACUCGGAAAACAUUUACGAGCCGCGCCCAAAUCGCAUCUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU .........(((((.(((((((((.....((....))....(((...(((((.(((((((((.......))).......))))))))))).))).....)))))).....))).))))). ( -37.01) >DroYak_CAF1 206671 120 - 1 CAGGGCAUUUGUCAAGCCGCUGUUAUCAAGGAAAAGCACUCGGAAAACAUAUACGAGCCGCGCCCAAAUCGCAUCUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU .........(((((.(((((((((......(((..((.((((...........))))..))(((((...........))))).........))).....)))))).....))).))))). ( -31.50) >consensus CAGGGCAUUUGUCAAGUCGCUGCUAUCAAGGAAAAGCACUCGGAAAACAUUUACGAGCCGAGCCCAAAACCCAUAUAUGGGCUCGAAAUGGUUCAAUAAAGCAGCUUUAUGGCGUGACAU .........(((((.(((((((((......(((.....((((...........)))).((((((((...........))))))))......))).....)))))).....))).))))). (-33.82 = -33.82 + 0.00)

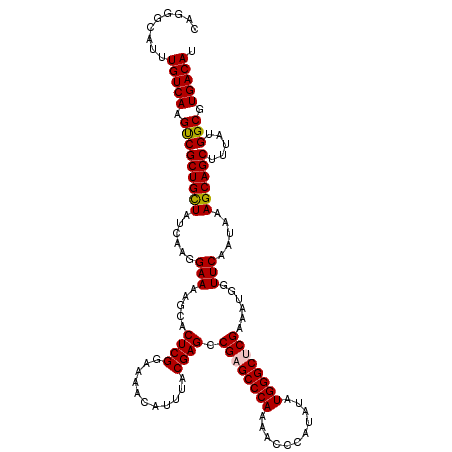

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:16 2006