| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 436,535 – 436,711 |
| Length | 176 |
| Max. P | 0.563881 |

| Location | 436,535 – 436,654 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -21.54 |
| Energy contribution | -23.58 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
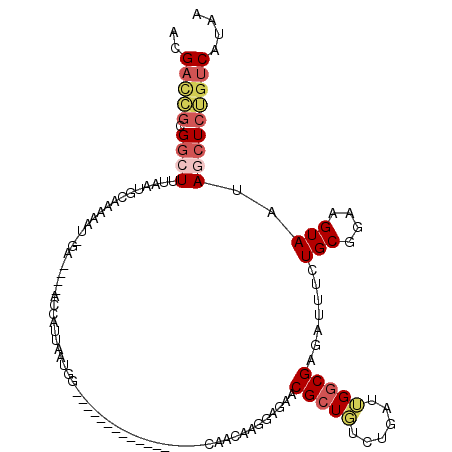
>2L_DroMel_CAF1 436535 119 + 22407834 GGGCUUGGAAUUGAAAAGAAGUGAUUGCAAGUAGUUUUCUGCGAAGUAGUCUAUAGUGAAGGUUUAUCGUC-UAAUAACUUUAUGACAGAGGUAUUACUUCCGCAGAAAUCUCGCCAAUC (((((((.(((((........))))).)))))((.(((((((((((((((((...((((((..(((.....-)))...))))))......)).))))))).)))))))).))..)).... ( -29.90) >DroSec_CAF1 13876 120 + 1 GGGUUCGAAAUUGAAGCGAAGUGAUUGCAGGGAGUUUUCUGCGAAGUAGUCUUUAGUGAAGAAGUAACAUCUUAAUAACUUUAUGACAGAGCUAUUACUUCCGCAGAAAUCUCGCCAAUC .(.((((....)))).).....(((((..(.(((.(((((((((((((((((((.((.(.(((((............))))).).))))))..))))))).)))))))).))).)))))) ( -32.50) >DroSim_CAF1 12988 120 + 1 GGGUUCGAAAUUGAAGCGAAGUGAUUGCAGGGAGUUUUCUGCGAAGUAGUCUUCAGUGAAGUCGUAUCGUCUUAAUAACUUUAUGACAGAGCUAUUACUUCCGCAGAAAUCUCGCCAAUC .(.((((....)))).).....(((((..(.(((.(((((((((((((((....(((...((((((..((.......))..))))))...)))))))))).)))))))).))).)))))) ( -34.90) >DroEre_CAF1 14001 120 + 1 CAGCUUGGAAUCCAAGAGAAAUGAUUGCAAGGAAGUUUCUGCAAAUUAGACUACAAUGGAGUUGUAUCGUCUUAACAACUUCAUGACCGAGAUGCUACUUCCGCAGAAAUCUCGCCAAUC ...((((.((((..........)))).))))((..(((((((.((.(((.((...((((((((((.........)))))))))).....))...))).))..)))))))..))....... ( -29.70) >DroYak_CAF1 13555 120 + 1 GAGUUUGGAAUUCAAGAGAAAUGAUUGCUAGGAUUUAUGUGCGAAGUAGUGUUUAAUGAAGUUUUAUCGUCUCAGUAACUUCAUGGCCGAGCUAUUACUUCCGCAGAUAUCUCGGCCAUC ((((.....)))).........(((.((..((((...((((.((((((((((((.((((((((....(......).))))))))....)))).))))))))))))...))))..)).))) ( -33.10) >consensus GGGUUUGGAAUUGAAGAGAAGUGAUUGCAAGGAGUUUUCUGCGAAGUAGUCUUUAGUGAAGUUGUAUCGUCUUAAUAACUUUAUGACAGAGCUAUUACUUCCGCAGAAAUCUCGCCAAUC ......................(((((....(((.((((((((((((((((((..((((((((((.........))))))))))....)))..))))))).)))))))).)))..))))) (-21.54 = -23.58 + 2.04)


| Location | 436,535 – 436,654 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 436535 119 - 22407834 GAUUGGCGAGAUUUCUGCGGAAGUAAUACCUCUGUCAUAAAGUUAUUA-GACGAUAAACCUUCACUAUAGACUACUUCGCAGAAAACUACUUGCAAUCACUUCUUUUCAAUUCCAAGCCC (((((.((((.((((((((.(((((........(((.(((.....)))-)))((.......)).........)))))))))))))....)))))))))...................... ( -24.80) >DroSec_CAF1 13876 120 - 1 GAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAAAGUUAUUAAGAUGUUACUUCUUCACUAAAGACUACUUCGCAGAAAACUCCCUGCAAUCACUUCGCUUCAAUUUCGAACCC (((((..(((.(((((((((((((((((..(((...(((....)))..)))))))))))).((......))......)))))))).)))....)))))...................... ( -26.90) >DroSim_CAF1 12988 120 - 1 GAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAAAGUUAUUAAGACGAUACGACUUCACUGAAGACUACUUCGCAGAAAACUCCCUGCAAUCACUUCGCUUCAAUUUCGAACCC (((((..(((.(((((((.(((((..((....((((.(((.....))).)))).))..)))))...((((....))))))))))).)))....)))))...................... ( -27.70) >DroEre_CAF1 14001 120 - 1 GAUUGGCGAGAUUUCUGCGGAAGUAGCAUCUCGGUCAUGAAGUUGUUAAGACGAUACAACUCCAUUGUAGUCUAAUUUGCAGAAACUUCCUUGCAAUCAUUUCUCUUGGAUUCCAAGCUG (((((.((((((..((((....)))).)))))).....((((((((..((((..(((((.....))))))))).....)))...)))))....)))))......(((((...)))))... ( -31.20) >DroYak_CAF1 13555 120 - 1 GAUGGCCGAGAUAUCUGCGGAAGUAAUAGCUCGGCCAUGAAGUUACUGAGACGAUAAAACUUCAUUAAACACUACUUCGCACAUAAAUCCUAGCAAUCAUUUCUCUUGAAUUCCAAACUC .(((((((((.(((.(((....)))))).)))))))))(((.(((..((((.((.......))...............((............)).......)))).))).)))....... ( -27.10) >consensus GAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAAAGUUAUUAAGACGAUACAACUUCACUAAAGACUACUUCGCAGAAAACUCCUUGCAAUCACUUCUCUUCAAUUCCAAACCC (((((..(((.((((((((((..((((((((.........)))))))).((.(......).))............)))))))))).)))....)))))...................... (-18.80 = -19.20 + 0.40)

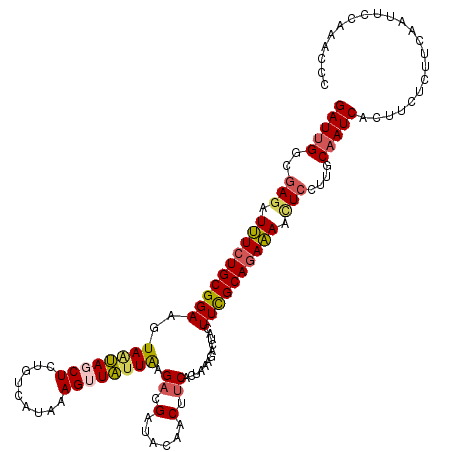
| Location | 436,614 – 436,711 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.20 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -11.50 |
| Energy contribution | -12.58 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 436614 97 - 22407834 ACGAACGCGGCUUUAAUGCAAAAAUU------GCCAUUAAUGG-----------------CAAGAAGGAGAACGCUGUCUGAUUGGCGAGAUUUCUGCGGAAGUAAUACCUCUGUCAUAA ......(((.......))).....((------((((....)))-----------------)))((.((((..(((((......))))).......(((....)))....)))).)).... ( -23.30) >DroSec_CAF1 13956 97 - 1 ACGACGGCGGCUUUAAUGCAAAAACU------ACCAUUAAUGG-----------------CAACAAGGAGAACGCUGUCUGAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAA ..(((((((((......)).....((------.((.....((.-----------------...)).))))..)))))))....(((((((.((..(((....)))..))))).))))... ( -26.70) >DroSim_CAF1 13068 103 - 1 ACGACGACGGCUUUAAUGCAAAAACCGACGAAACCAUUAAUGG-----------------CAACAAGGAGAACGCUGUCUGAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAA ..(((((((((.((((((................))))))((.-----------------...))........))))))........(((.((..(((....)))..))))).))).... ( -22.49) >DroEre_CAF1 14081 117 - 1 ACGAUCGUGACUGUAAUGCAA-AAU-GA-UUUACCAUUAAUCGGAACAAGCUUUCUCGUACAACAAACAGAACGCUUUUUGAUUGGCGAGAUUUCUGCGGAAGUAGCAUCUCGGUCAUGA ....((((((((.....((..-(((-(.-.....))))((((((((..(((.((((.((.......)))))).))))))))))).))(((((..((((....)))).))))))))))))) ( -34.80) >DroYak_CAF1 13635 118 - 1 ACGAUCGUGGCUUUAUUGCAAAAAU-GA-UAUACCAUUAAUCGGAAUAAGCUUCCUUGUUCAACAAAUAGAACGCUUUUUGAUGGCCGAGAUAUCUGCGGAAGUAAUAGCUCGGCCAUGA .((((.((((..(((((.....)))-))-....))))..))))(((((((....)))))))..((((.((....)).))))(((((((((.(((.(((....)))))).))))))))).. ( -37.10) >consensus ACGACCGCGGCUUUAAUGCAAAAAU_GA____ACCAUUAAUGG_________________CAACAAGGAGAACGCUGUCUGAUUGGCGAGAUUUCUGCGGAAGUAAUAGCUCUGUCAUAA ..(((((.((((............................................................(((((......))))).......(((....)))..))))))))).... (-11.50 = -12.58 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:31 2006