| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,019,906 – 4,020,007 |
| Length | 101 |
| Max. P | 0.905772 |

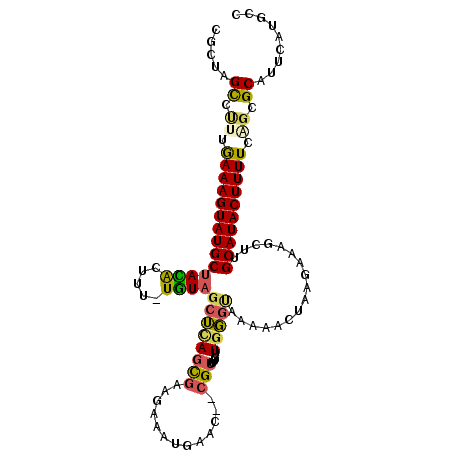
| Location | 4,019,906 – 4,020,007 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.45 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
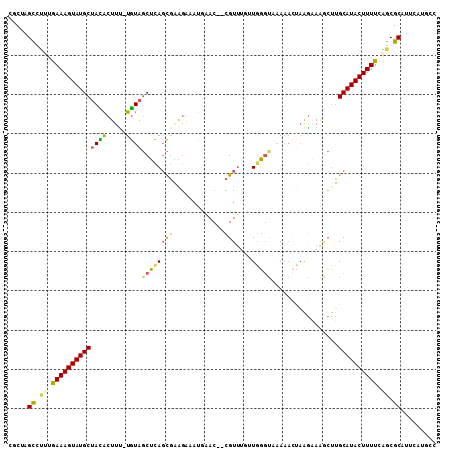
>2L_DroMel_CAF1 4019906 101 - 22407834 CGCUAGCCCUUGAAAGUAUGCUACACUUU-UGUAGCUCAGCGAAGAAAUGAAC--CGUUUGUUCGGUUAA-----GGUAAGCUUGCAUACUUUUCAGCGCAUUCAUGCC .....((.((.((((((((((((((....-))))))...((.(((.....(((--((......)))))..-----......))))))))))))).)).))......... ( -26.52) >DroVir_CAF1 230862 104 - 1 GGC-AGCAGCUAAAAGUAUGCAAUGCUUUUUAUG-CGUACUUUGUGUCUUGGCUAUGUUUG-UGUGUAUAAAUCAAAUAACAUGGCAUACUUUUGCGCGCGUUGAUU-- ..(-(((.(((((((((((((.(((...((((((-(((((..(((((....)).)))...)-))))))))))........))).))))))))))).))..))))...-- ( -27.90) >DroSec_CAF1 153397 106 - 1 CGCUAGCCUUUGAAAGUAUGCUACACUUU-UGUAGCUCAGCGAAGAAAUGAAC--CGUUUGUUGGGUAAAAACUAAGAAGGCUUGCAUACUUUUCAGCGCAUUCAUGCC .((.(((((((...(((..((((((....-))))))(((((((((........--).))))))))......)))..))))))).))............(((....))). ( -28.10) >DroSim_CAF1 157788 106 - 1 CGCUAGUUUAUAAAAGUAUGCUACACUUC-UGUAGCUCAGCGAAGAAAUUAAC--CGUUUUAUGGGCUAAAACUAAGAAAGCUUGCAUACUUUUCAGCGCAUUCAUGCC .(((.(.....((((((((((.....(((-(.((((((((((...........--)))....)))))))......)))).....))))))))))))))(((....))). ( -24.20) >DroEre_CAF1 159890 102 - 1 CACAAGCCUUGGAAAGUAUGCUAUGCUUU-UGUAGCUCAGCGGAGAAACAAAC--CGU----UGGGUAAAAACUAAGAAGACUUGCAUACUUUUCGGCGCACACAUCUC .....((.(((((((((((((....((((-(...(((((((((.........)--)))----)))))........)))))....))))))))))))).))......... ( -33.60) >DroYak_CAF1 164190 106 - 1 CACAAGCCUUUGAAAGUAUGCUACACUUU-UGUAGCUCAGCGAAGUUAAGAAC--CGUUAGUUGGCUGAAAACUAAGAAAGCCUGCAUACUUUUCGGCGCAUUCAUCGC .....(((...((((((((((....((((-(.(((.(((((.((.((((....--..)))))).)))))...))).)))))...)))))))))).)))........... ( -29.10) >consensus CGCUAGCCUUUGAAAGUAUGCUACACUUU_UGUAGCUCAGCGAAGAAAUGAAC__CGUUUGUUGGGUAAAAACUAAGAAAGCUUGCAUACUUUUCAGCGCAUUCAUGCC .....((.((.((((((((((((((.....))))((((((((.............)))....))))).................)))))))))).)).))......... (-13.75 = -13.45 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:01 2006