| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,011,257 – 4,011,373 |
| Length | 116 |
| Max. P | 0.500000 |

| Location | 4,011,257 – 4,011,373 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
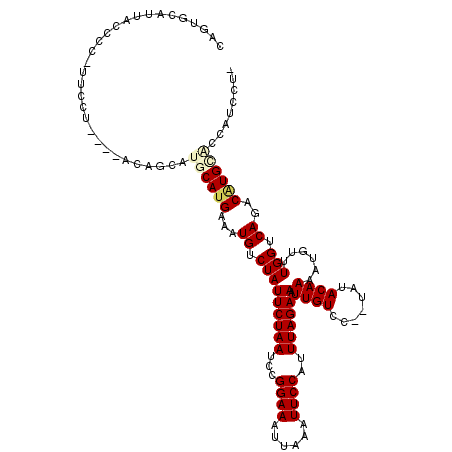
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
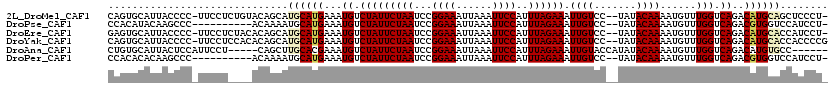
>2L_DroMel_CAF1 4011257 116 + 22407834 CAGUGCAUUACCCC-UUCCUCUGUACAGCAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC--UAUACAAAAUGUUUGGUCAGACAUGCAGCUCCCU- ..(((((.......-......)))))(((.((((((.........((((((...((((......))))..)))))).(((.((--...((.....))..)).))).)))))))))....- ( -26.22) >DroPse_CAF1 223725 107 + 1 CCACAUACAAGCCC----------ACAAAAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC--UAUACAAAAUGUUUGGUCAGACGUGGUCCAUCCU- ((((...((.((..----------.......)).))....((((.((((((...((((......))))..))))))...(.((--...((.....))..)).)))))))))........- ( -20.20) >DroEre_CAF1 150230 116 + 1 GAGUGCAUUACCCC-UUCCUCUACACAGCAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC--UAUACAAAAUGUUUGGUCAGACAUGCACCAUCCU- (.((((........-..........((.......))..((((((.((((((...((((......))))..))))))...(.((--...((.....))..)).)))))))))))).....- ( -21.10) >DroYak_CAF1 155401 117 + 1 CAGUGCAUUACCCC-UUCCUCCACACAGCAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC--UAUACAAAAUGUUUGGUCAGACAUGCACCACCCCG ..((((........-..........((.......))..((((((.((((((...((((......))))..))))))...(.((--...((.....))..)).)))))))))))....... ( -20.90) >DroAna_CAF1 169471 109 + 1 CUGUGCAUUACUCCAUUCCU-----CAGCUUGCACGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUACCAUAUACAAAAUGUUUGGUCAGACAUGUGCC------ .((((((.............-----.....))))))..((((((.((((((...((((......))))..))))))...(.((((...((.....)).))))))))))).....------ ( -22.87) >DroPer_CAF1 224180 107 + 1 CCACACACAAGCCC----------ACAAAAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC--UAUACAAAAUGUUUGGUCAGACGUGGUCCAUCCU- ((((...((.((..----------.......)).))....((((.((((((...((((......))))..))))))...(.((--...((.....))..)).)))))))))........- ( -20.20) >consensus CAGUGCAUUACCCC_UUCCU____ACAGCAUGCAUGAAAUGUCUAUUCUAAUCCGGAAAUUAAAUUCCAUUUAGAAAUUGUCC__UAUACAAAAUGUUUGGUCAGACAUGCACCAUCCU_ ..............................((((((...((.(((((((((...((((......))))..)))))).((((.......))))......))).))..))))))........ (-14.40 = -14.90 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:59 2006