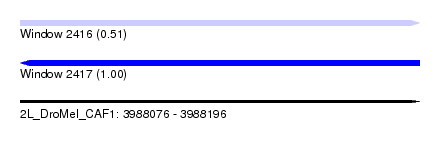
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,988,076 – 3,988,196 |
| Length | 120 |
| Max. P | 0.995429 |

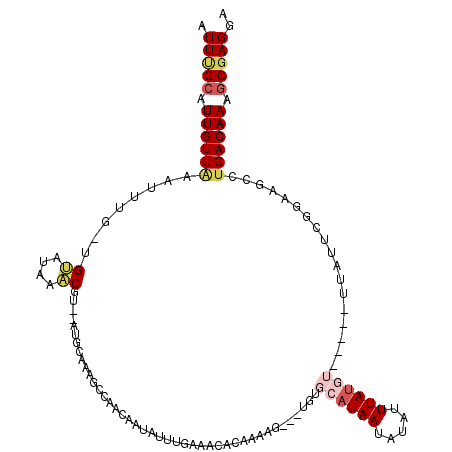
| Location | 3,988,076 – 3,988,196 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.04 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -2.95 |
| Energy contribution | -3.38 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.16 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3988076 120 + 22407834 UCCUGGCUUUGUCAGGCUUGCGAAAAAAAACAACAUAAAUAUAUUAUGCACAAAACUUUUGUGCUUCAAAUAUUGUUGCCUUUGCAUACACGUUUGUACAUCAAAUUUAACAAUGCAAAU .((((((...)))))).((((..........................((((((.....)))))).......(((((((..((((..((((....))))...))))..))))))))))).. ( -24.30) >DroSec_CAF1 121159 111 + 1 UUCUCGCUUUGUCAGGCUCCCGAAUAA-----ACAUAAAUAUAUUAUGCACA--UCUUUUGUGUUUCAAAUAUUCUUGCCUUUGCAU--ACGUUUAUACACCAAAUUUAACAAUGCAAAU .....((.((((.((((....(((((.-----.(((((.....)))))((((--.....)))).......)))))..))))......--..((....))..........)))).)).... ( -16.00) >DroSim_CAF1 125747 100 + 1 UUCUCGCUUUGUCAGGCUCGCGAAUAA-----ACAUAAAUAUAUUAUGCACA--UCUUUUGUGCUUCAAAUAUU-----------GU--ACGUUUAUACACCAAAUUUAACAAUGCAAAU .....((.((((.(((...(.(.((((-----((.((...(((((..(((((--.....)))))....))))).-----------.)--).)))))).).)....))).)))).)).... ( -16.20) >DroEre_CAF1 126333 94 + 1 UCCUCGCUUUGUCCGCUUACUGAAUAA-----AUAUAAAUAUAUUAU---------------GUUUCGAAUGCUGUUGGCUUUGCAU--UCGUUUAUUCA----UUUCAACAAUGCGAAU ...((((.((((........(((((((-----((((((.....))))---------------))..(((((((..........))))--))).)))))))----.....)))).)))).. ( -20.42) >DroYak_CAF1 129756 92 + 1 UCCUCGUUUUGUCAGGUUUCUGAAUAA-----ACAUAAAUAUAUUA-----------------UUGCGAAUGCUGUUGGCUUUGCAU--ACGAAUAUUCA----UUUCAACAAUGCAAAU .....((((..((((....))))..))-----))......(((((.-----------------.((((((.((.....)))))))).--...)))))...----................ ( -14.30) >consensus UCCUCGCUUUGUCAGGCUCCCGAAUAA_____ACAUAAAUAUAUUAUGCACA___CUUUUGUGUUUCAAAUAUUGUUGCCUUUGCAU__ACGUUUAUACA_CAAAUUUAACAAUGCAAAU .....((.((((.............................((((..(((((.......)))))....))))...................((....))..........)))).)).... ( -2.95 = -3.38 + 0.43)

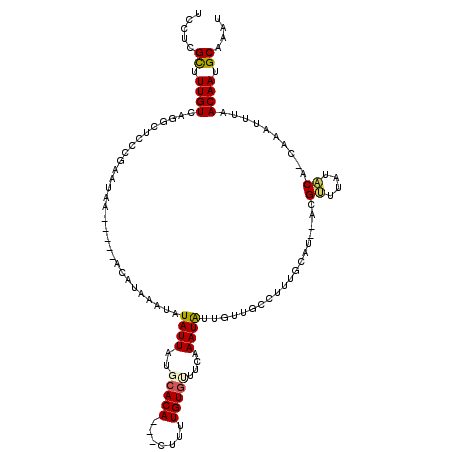
| Location | 3,988,076 – 3,988,196 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.04 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -5.86 |
| Energy contribution | -7.18 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.24 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3988076 120 - 22407834 AUUUGCAUUGUUAAAUUUGAUGUACAAACGUGUAUGCAAAGGCAACAAUAUUUGAAGCACAAAAGUUUUGUGCAUAAUAUAUUUAUGUUGUUUUUUUUCGCAAGCCUGACAAAGCCAGGA .(((((((((((...((((.((((((....))))))))))...))))))....(((((((((.....))))))(((((((....))))))).....))))))))((((.......)))). ( -28.30) >DroSec_CAF1 121159 111 - 1 AUUUGCAUUGUUAAAUUUGGUGUAUAAACGU--AUGCAAAGGCAAGAAUAUUUGAAACACAAAAGA--UGUGCAUAAUAUAUUUAUGU-----UUAUUCGGGAGCCUGACAAAGCGAGAA .(((((.(((((........((((((....)--))))).((((..(((((.......((((.....--))))(((((.....))))).-----.)))))....))))))))).))))).. ( -24.20) >DroSim_CAF1 125747 100 - 1 AUUUGCAUUGUUAAAUUUGGUGUAUAAACGU--AC-----------AAUAUUUGAAGCACAAAAGA--UGUGCAUAAUAUAUUUAUGU-----UUAUUCGCGAGCCUGACAAAGCGAGAA .(((((.((((((...((.(((.((((((((--(.-----------(((((.....(((((.....--))))).....))))))))))-----)))).))).))..)))))).))))).. ( -28.40) >DroEre_CAF1 126333 94 - 1 AUUCGCAUUGUUGAAA----UGAAUAAACGA--AUGCAAAGCCAACAGCAUUCGAAAC---------------AUAAUAUAUUUAUAU-----UUAUUCAGUAAGCGGACAAAGCGAGGA .(((((.((((((...----(((((((((((--((((..........)))))))....---------------((((.....)))).)-----))))))).....).))))).))))).. ( -24.10) >DroYak_CAF1 129756 92 - 1 AUUUGCAUUGUUGAAA----UGAAUAUUCGU--AUGCAAAGCCAACAGCAUUCGCAA-----------------UAAUAUAUUUAUGU-----UUAUUCAGAAACCUGACAAAACGAGGA .(((((((...((((.----......)))).--))))))).((....((....))..-----------------...........(((-----((..((((....))))..))))).)). ( -16.00) >consensus AUUUGCAUUGUUAAAUUUG_UGUAUAAACGU__AUGCAAAGCCAACAAUAUUUGAAACACAAAAG___UGUGCAUAAUAUAUUUAUGU_____UUAUUCGGAAGCCUGACAAAGCGAGGA .(((((.((((((........((....))..........................................((((((.....))))))..................)))))).))))).. ( -5.86 = -7.18 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:51 2006