
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,980,733 – 3,980,831 |
| Length | 98 |
| Max. P | 0.975687 |

| Location | 3,980,733 – 3,980,831 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3980733 98 + 22407834 UCCGUCCGU-UGGGUAACAAUUCUCUGCAGGUGGCAGGAAAAAACUGGGAAAAUGUCCCACGAGAGAUGGCCACUCAAAGAUACCCUGGAAAUUAGAAA ....((((.-.(((((.......(((...((((((..........(((((.....)))))(....)...))))))...))))))))))))......... ( -27.31) >DroSec_CAF1 113823 98 + 1 UCCGUCCGU-UGGGUAACAAUUCUCUGCAGGUGGCAGGGAAACACUGGGAAAAUGUCCCAGGAGAGAUGGCCACUCAGAGAUACCCUGGAAAUUAGAAA ....((((.-.(((((.....((((((..((((((..(....).((((((.....))))))........)))))))))))))))))))))......... ( -38.60) >DroSim_CAF1 118341 98 + 1 UCCGUCCGU-UGGGUAACAAUUCUCUGCAGGUGGCAGGGAAACACUGGGAAAAUGUCCCAGGAGAGAUGGCCACUCAGAGAUACCCUGGAAAUAAGAAA ....((((.-.(((((.....((((((..((((((..(....).((((((.....))))))........)))))))))))))))))))))......... ( -38.60) >DroEre_CAF1 119030 98 + 1 UCCGUCCGU-UGGGUAGCAAUUCUCUGCUGGUGGCAGGGAAAAACUGGGAAAAUGUCCCCGGAGAGAUGGCCACUCAGAGAUACCCUGGAAAUUAGAAA ....((((.-.(((((.....((((((..((((((.........(((((........))))).......)))))))))))))))))))))......... ( -33.39) >DroYak_CAF1 122371 98 + 1 UCCGUCCGU-UGGGUAGAAAUUCUCUGCAGGUGGCAGGGAAAAACUGGGAAAAUGUCCCAGGAGAGAUGGCCACUUGGAGAUACCCUGGAAAUUAGAAA ....((((.-.(((((.......(((.((((((((.........((((((.....))))))........)))))))).))))))))))))......... ( -36.64) >DroAna_CAF1 135112 98 + 1 UCCGGCCGAAUGGAAAACAAAUCUGUGCAGUUGGCAGGCUAA-AUGGUGAAAAGGUCCUCGGAGAGAUGGCCACUUAGAGAUACCCUGAAAAUUAGAAG .((.(((((...(...(((....))).)..))))).))((((-..........((((.((.....)).))))..((((.......))))...))))... ( -19.10) >consensus UCCGUCCGU_UGGGUAACAAUUCUCUGCAGGUGGCAGGGAAAAACUGGGAAAAUGUCCCAGGAGAGAUGGCCACUCAGAGAUACCCUGGAAAUUAGAAA ....((((...(((((.....((((((..((((((.........((((((.....))))))........)))))))))))))))))))))......... (-27.74 = -28.58 + 0.84)

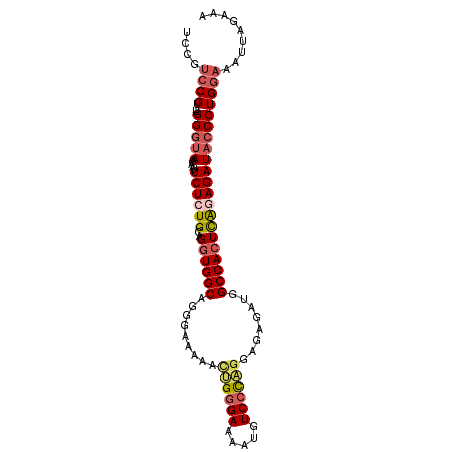

| Location | 3,980,733 – 3,980,831 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -22.92 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3980733 98 - 22407834 UUUCUAAUUUCCAGGGUAUCUUUGAGUGGCCAUCUCUCGUGGGACAUUUUCCCAGUUUUUUCCUGCCACCUGCAGAGAAUUGUUACCCA-ACGGACGGA .........(((.(((((((((((.(((((.........(((((.....)))))..........)))))...)))))).....))))).-..))).... ( -27.51) >DroSec_CAF1 113823 98 - 1 UUUCUAAUUUCCAGGGUAUCUCUGAGUGGCCAUCUCUCCUGGGACAUUUUCCCAGUGUUUCCCUGCCACCUGCAGAGAAUUGUUACCCA-ACGGACGGA .........(((.(((((((((((.(((((.......(((((((.....)))))).).......)))))...)))))).....))))).-..))).... ( -33.94) >DroSim_CAF1 118341 98 - 1 UUUCUUAUUUCCAGGGUAUCUCUGAGUGGCCAUCUCUCCUGGGACAUUUUCCCAGUGUUUCCCUGCCACCUGCAGAGAAUUGUUACCCA-ACGGACGGA .........(((.(((((((((((.(((((.......(((((((.....)))))).).......)))))...)))))).....))))).-..))).... ( -33.94) >DroEre_CAF1 119030 98 - 1 UUUCUAAUUUCCAGGGUAUCUCUGAGUGGCCAUCUCUCCGGGGACAUUUUCCCAGUUUUUCCCUGCCACCAGCAGAGAAUUGCUACCCA-ACGGACGGA .........(((.(((((((((((.(((((..........((((.....))))...........)))))...)))))).....))))).-..))).... ( -29.70) >DroYak_CAF1 122371 98 - 1 UUUCUAAUUUCCAGGGUAUCUCCAAGUGGCCAUCUCUCCUGGGACAUUUUCCCAGUUUUUCCCUGCCACCUGCAGAGAAUUUCUACCCA-ACGGACGGA .........(((.(((((((((((.(((((........((((((.....)))))).........))))).))..)))).....))))).-..))).... ( -28.63) >DroAna_CAF1 135112 98 - 1 CUUCUAAUUUUCAGGGUAUCUCUAAGUGGCCAUCUCUCCGAGGACCUUUUCACCAU-UUAGCCUGCCAACUGCACAGAUUUGUUUUCCAUUCGGCCGGA ..............((((...((((((((.......((....))........))))-))))..))))..(.((...((..((.....)).)).)).).. ( -14.26) >consensus UUUCUAAUUUCCAGGGUAUCUCUGAGUGGCCAUCUCUCCUGGGACAUUUUCCCAGUUUUUCCCUGCCACCUGCAGAGAAUUGUUACCCA_ACGGACGGA .............(((((((((((.(((((........((((((.....)))))).........)))))...)))))).....)))))........... (-22.92 = -24.28 + 1.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:49 2006