
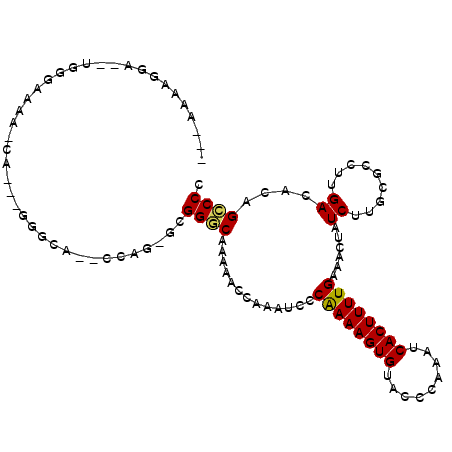
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,955,830 – 3,955,933 |
| Length | 103 |
| Max. P | 0.929998 |

| Location | 3,955,830 – 3,955,933 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
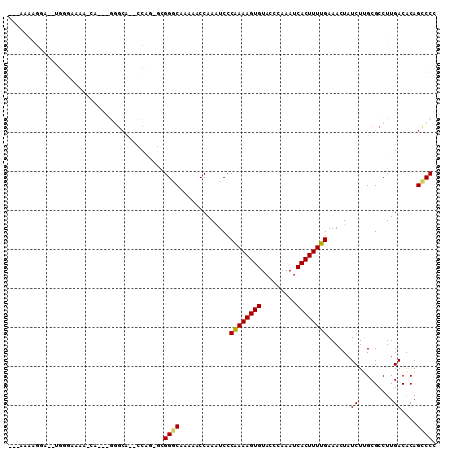
>2L_DroMel_CAF1 3955830 103 + 22407834 ---AAAGGGA--UGGGAAAA-CACAAGGGCAAACCAG-GCGGGCAAAAACCAAAUCCCGAAAGUGUACCCAAAUCACUUUUGAAACUAUCUUGCGCCUUGACACAGCCCC ---...(((.--(((.....-).(((((((((...((-..((.......))......((((((((.........))))))))...))...)))).)))))...)).))). ( -25.30) >DroGri_CAF1 134438 86 + 1 ---------------CACAG------CAGCA--GCUC-CAGGACAAAAACCAAAUCCCAAAAGUGUACCCAAAUCACUUUUGAAACUAUCUGGCGCCUUGACGCAGUCCC ---------------...((------(....--))).-..((((.............((((((((.........))))))))..........(((......))).)))). ( -18.40) >DroSec_CAF1 88896 102 + 1 ---A-AAGGA--UGGGAAAA-CACAAGGGCAACCCAG-GCGGGCAAAAACCAAAUCCCGAAAGUGUACCCAAAUCACUUUUGAAACUAUCUUGCGCCUUGACACAGCCCC ---.-.....--.(((....-..((((((((((((..-..)))..............((((((((.........))))))))........)))).)))))......))). ( -26.40) >DroSim_CAF1 88108 103 + 1 ---AAAAGGA--UGGGAAAA-CACAAGGGCAGCCCAG-GCGGGCAAAAACCAAAUCCCGAAAGUGUACCCAAAUCACUUUUGAAACUAUCUUGCGCCUUGACACAGCCCC ---.......--.(((....-..((((((((((((..-..)))).............((((((((.........)))))))).........))).)))))......))). ( -30.30) >DroMoj_CAF1 152625 100 + 1 ----ACAGCAGCUCCAGCAGCAC---CAGCA--GCAGCCAGGACAA-AACCAAAUCCCAAAAGUGUACCCAAAUCACUUUUGAAACUAUCUGGCGCCUUGACGCAGUCCG ----...((.((....)).((..---..)).--)).....((((..-..........((((((((.........))))))))..........(((......))).)))). ( -23.20) >DroAna_CAF1 109377 99 + 1 GAGGAAAGGA--GGGGCAA-------GG-AGGUGCUU-UUGGGCAAAAACCAAGUCCCAAAAGUGUACCCAAAUCACUUUUGAAACUAUCUUGCGCCUUGACACAGACCC ........((--((.((((-------((-.(((((((-((((((.........).))))))))).....((((.....))))..))).)))))).))))........... ( -29.40) >consensus ___AAAAGGA__UGGGAAAA_CA___GGGCA__CCAG_GCGGGCAAAAACCAAAUCCCAAAAGUGUACCCAAAUCACUUUUGAAACUAUCUUGCGCCUUGACACAGCCCC ........................................((((.............((((((((.........))))))))......((.........))....)))). (-12.92 = -12.50 + -0.42)

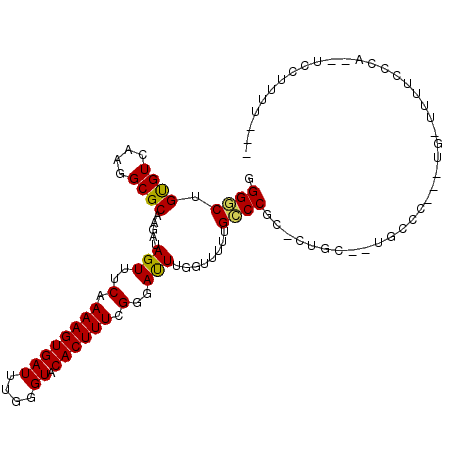
| Location | 3,955,830 – 3,955,933 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -19.21 |
| Energy contribution | -18.68 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
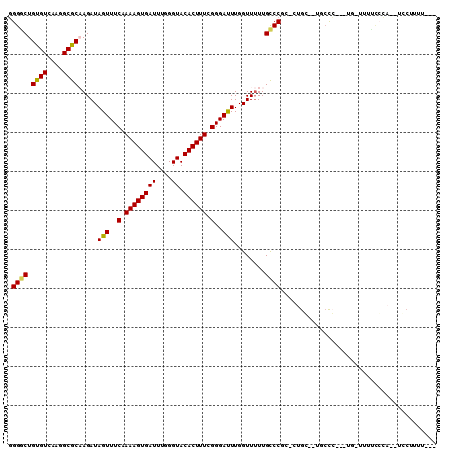
>2L_DroMel_CAF1 3955830 103 - 22407834 GGGGCUGUGUCAAGGCGCAAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUCGGGAUUUGGUUUUUGCCCGC-CUGGUUUGCCCUUGUG-UUUUCCCA--UCCCUUU--- ((((..(((..((((((((((..(((..(.((((((((....)).)))))).)..))).(((....(((...-..)))..)))))))))-))))..))--)))))..--- ( -29.90) >DroGri_CAF1 134438 86 - 1 GGGACUGCGUCAAGGCGCCAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUUGGGAUUUGGUUUUUGUCCUG-GAGC--UGCUG------CUGUG--------------- (((((.((((....)))).....(((..((((((((((....)).))))))))..)))........)))))(-.(((--....)------)).).--------------- ( -26.20) >DroSec_CAF1 88896 102 - 1 GGGGCUGUGUCAAGGCGCAAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUCGGGAUUUGGUUUUUGCCCGC-CUGGGUUGCCCUUGUG-UUUUCCCA--UCCUU-U--- ((((..(((..((((((((((..(((..(.((((((((....)).)))))).)..))).(((....((((..-..)))).)))))))))-))))..))--)))))-.--- ( -30.40) >DroSim_CAF1 88108 103 - 1 GGGGCUGUGUCAAGGCGCAAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUCGGGAUUUGGUUUUUGCCCGC-CUGGGCUGCCCUUGUG-UUUUCCCA--UCCUUUU--- ((((..(((..((((((((((..(((..(.((((((((....)).)))))).)..))).(((....((((..-..)))).)))))))))-))))..))--)))))..--- ( -32.00) >DroMoj_CAF1 152625 100 - 1 CGGACUGCGUCAAGGCGCCAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUUGGGAUUUGGUU-UUGUCCUGGCUGC--UGCUG---GUGCUGCUGGAGCUGCUGU---- (((...((..((.((((((((.((((..((((((((((....)).))))))))((((......-..)))).....))--)))))---)))))..))..))..))).---- ( -35.80) >DroAna_CAF1 109377 99 - 1 GGGUCUGUGUCAAGGCGCAAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUUGGGACUUGGUUUUUGCCCAA-AAGCACCU-CC-------UUGCCCC--UCCUUUCCUC ((((..((((...((.((((((.(((..((((((((((....)).))))))))..)))....))))))))..-..))))..-..-------..)))).--.......... ( -30.10) >consensus GGGGCUGUGUCAAGGCGCAAGAUAGUUUCAAAAGUGAUUUGGGUACACUUUCGGGAUUUGGUUUUUGCCCGC_CUGC__UGCCC___UG_UUUUCCCA__UCCUUUU___ .((((.((((....)))).....(((..(.((((((((....)).)))))).)..)))........))))........................................ (-19.21 = -18.68 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:41 2006