
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,933,043 – 3,933,135 |
| Length | 92 |
| Max. P | 0.867397 |

| Location | 3,933,043 – 3,933,135 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
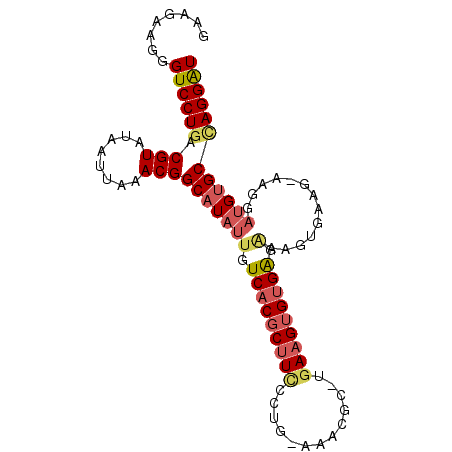
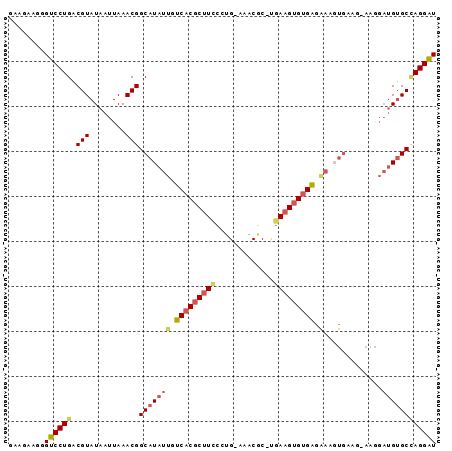
>2L_DroMel_CAF1 3933043 92 + 22407834 GAAGAAGGGUCCUGGCGUAUAAUUAAACGGCAUAUUGUCACGCUUCCCUG-AAACGC-AGAAGUGUGAGAAAGUGA-CAAAGGAUGUGCCAGGAU ........((((((((((((..........(((.((.(((((((((..((-.....)-)))))))))).)).))).-......)))))))))))) ( -30.63) >DroSec_CAF1 66293 93 + 1 GAAGAAGGGUCCUGGCGUAUAAUUAAACGGCAUAUUGUCACGCUUCCCUG-ACACAC-UGAAGUGUGAGAAAGUGAAGAAAGGAUGUGCCAGGAU ........((((((((((((..((...(..(((.((.(((((((((..((-...)).-.))))))))).)).)))..).))..)))))))))))) ( -31.30) >DroSim_CAF1 65180 93 + 1 GAAGAAGGGUCCUGGCGUAUAAUUAAACGGCAUAUUGUCACGCUUCCCUG-AAACGC-UGAAGUGUGAGAAAGUGAAGAAAGGAUGUGCCAGGAU ........((((((((((((..((...(..(((.((.(((((((((....-......-.))))))))).)).)))..).))..)))))))))))) ( -30.90) >DroEre_CAF1 70696 92 + 1 GACGAAAGGUCCUGACGUAUAAUUAAACGGCAUAUUGUCACGCUUUCGUG-AAACGC-UGAAGUGUGAGGAGCUGAAG-AGGGAUGUGCUAGGAU ..(....)((((((.(((((..((...((((...((.(((((((((.(((-...)))-.))))))))).))))))...-))..))))).)))))) ( -28.00) >DroYak_CAF1 73190 81 + 1 GCAGAAGGGUCCUGACGUAUAAUUAAACGGCAUAUUGUCACGCUUUCCUGCAAACGC-UGAAGUGUGAG-------------GAUGUGCCAGGAU ........((((((.(((........)))(((((((.(((((((((...((....))-.))))))))).-------------))))))))))))) ( -32.30) >DroPer_CAF1 101784 84 + 1 ---GAAAGGUCCUGACGUAUAAUUAAACGGCAUAUUGUCACGCAUCCCUG-AAGCUCCCAAAGAGAGGGAAAGUGAAA-------GAGCAAGGGU ---......((((..(((........)))((......((((...(((((.-...(((.....))))))))..))))..-------..)).)))). ( -18.10) >consensus GAAGAAGGGUCCUGACGUAUAAUUAAACGGCAUAUUGUCACGCUUCCCUG_AAACGC_UGAAGUGUGAGAAAGUGAAG_AAGGAUGUGCCAGGAU ........((((((.(((........)))(((((((.(((((((((.............))))))))).).............)))))))))))) (-16.61 = -17.45 + 0.84)

| Location | 3,933,043 – 3,933,135 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -13.40 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
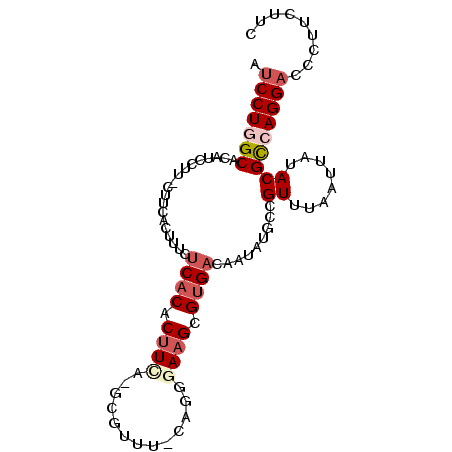
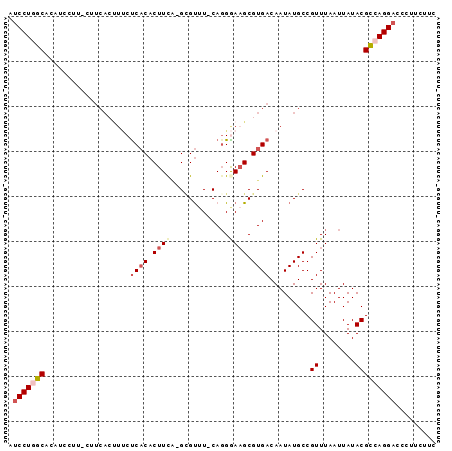
>2L_DroMel_CAF1 3933043 92 - 22407834 AUCCUGGCACAUCCUUUG-UCACUUUCUCACACUUCU-GCGUUU-CAGGGAAGCGUGACAAUAUGCCGUUUAAUUAUACGCCAGGACCCUUCUUC .(((((((.(((...(((-((((((((((((.(....-).))..-..)))))).))))))).)))..((........)))))))))......... ( -27.00) >DroSec_CAF1 66293 93 - 1 AUCCUGGCACAUCCUUUCUUCACUUUCUCACACUUCA-GUGUGU-CAGGGAAGCGUGACAAUAUGCCGUUUAAUUAUACGCCAGGACCCUUCUUC .(((((((...........((((((((((((((....-..))))-..)))))).)))).........((........)))))))))......... ( -26.20) >DroSim_CAF1 65180 93 - 1 AUCCUGGCACAUCCUUUCUUCACUUUCUCACACUUCA-GCGUUU-CAGGGAAGCGUGACAAUAUGCCGUUUAAUUAUACGCCAGGACCCUUCUUC .(((((((............((..((.((((.((((.-.(....-..).)))).)))).))..))..((........)))))))))......... ( -22.60) >DroEre_CAF1 70696 92 - 1 AUCCUAGCACAUCCCU-CUUCAGCUCCUCACACUUCA-GCGUUU-CACGAAAGCGUGACAAUAUGCCGUUUAAUUAUACGUCAGGACCUUUCGUC ................-....((.((((.((......-((((.(-((((....)))))....)))).((........)))).)))).))...... ( -14.00) >DroYak_CAF1 73190 81 - 1 AUCCUGGCACAUC-------------CUCACACUUCA-GCGUUUGCAGGAAAGCGUGACAAUAUGCCGUUUAAUUAUACGUCAGGACCCUUCUGC .(((((((...((-------------((((.((....-..)).)).))))..(((((....))))).............)))))))......... ( -19.60) >DroPer_CAF1 101784 84 - 1 ACCCUUGCUC-------UUUCACUUUCCCUCUCUUUGGGAGCUU-CAGGGAUGCGUGACAAUAUGCCGUUUAAUUAUACGUCAGGACCUUUC--- ..(((.((..-------..((((..((((((((.....)))...-.)))))...)))).........((........)))).))).......--- ( -16.20) >consensus AUCCUGGCACAUCCUU_CUUCACUUUCUCACACUUCA_GCGUUU_CAGGGAAGCGUGACAAUAUGCCGUUUAAUUAUACGCCAGGACCCUUCUUC .(((((((...................((((.((((.............)))).)))).........((........)))))))))......... (-13.40 = -14.40 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:35 2006