
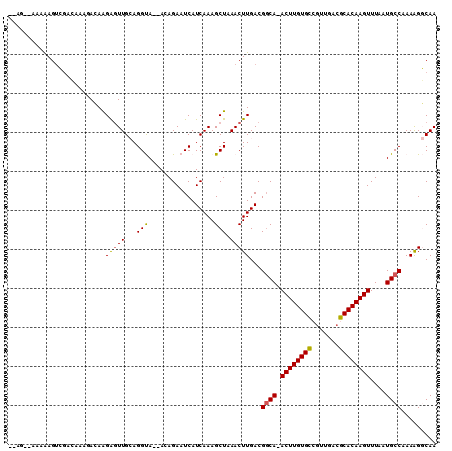
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,898,139 – 3,898,265 |
| Length | 126 |
| Max. P | 0.717640 |

| Location | 3,898,139 – 3,898,240 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3898139 101 + 22407834 ------AAAAAGUCGACAAAGACAGGAGUUGCAGGUA--AGAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGCCACUGACGCACAAGUUUAAUGCCAAAGGGCAA ------.....(((......)))....(((((((((.--..((...........))..))))).))))(-((((((((.......)))))))))...((((....)))). ( -26.40) >DroPse_CAF1 52686 107 + 1 AUAGGUUAAGAGUGGAGAC--AGAAGAGUUGAAGGCAGAACAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGUAGUUGACGCACAAGUUUAAUGCCAAAAGCCAA ....((((((..((....)--)..((..((((.((..........)).))))..))...))))))((((-((((((((.......))))))))....))))......... ( -25.80) >DroEre_CAF1 30098 105 + 1 CCAG--AAAAAGUCGCCAAAGACAGGAGUUGCAGGUA--AGAGAAUCAUCAAAGCUAAACUUGACGUCA-ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAGGGCAA ....--........(((...(((....)))...((((--.((......)).....((((((((.(((((-((.......)))))))..)))))))).))))....))).. ( -26.80) >DroYak_CAF1 32493 101 + 1 ------AAAAAGUCGGCAAAGACAGGAGUUGCAGGUA--AGAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAGGGCAA ------.....(((......)))....(((((((((.--..((...........))..))))).))))(-((((((((.......)))))))))...((((....)))). ( -26.40) >DroAna_CAF1 45997 101 + 1 GCAA--AAAGAG-----AUAUAGAAGGCUUGCAGGUA--ACAGAAUCAUCAAAGCUAAACUUGACGGCAAACUUGUGUCGUUGACGCACAAGUUUAAUGCCAAAAGGCAA ((..--......-----........(((((...(((.--.....)))....)))))...(((...((((((((((((.((....))))))))))...))))..))))).. ( -25.20) >DroPer_CAF1 53091 107 + 1 AUAGGUUAAGAGUGGAGAC--AGAAGAGUUGAAGGCAGAACAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGUAGUUGACGCACAAGUUUAAUGCCAAAAGCCAA ....((((((..((....)--)..((..((((.((..........)).))))..))...))))))((((-((((((((.......))))))))....))))......... ( -25.80) >consensus __AG__AAAAAGUCGACAAAGACAAGAGUUGCAGGUA__ACAGAAUCAUCAAAGCUAAACUUGACGGCA_ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAAGGCAA .........................(((((...(((.................))).)))))...((((.((((((((.......))))))))....))))......... (-17.51 = -17.23 + -0.28)


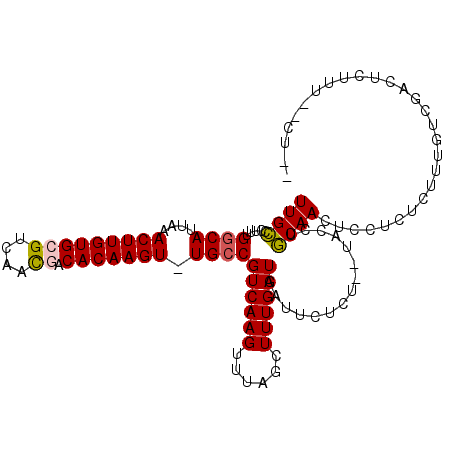
| Location | 3,898,139 – 3,898,240 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3898139 101 - 22407834 UUGCCCUUUGGCAUUAAACUUGUGCGUCAGUGGCACAAGU-UGCCGUCAAGUUUAGCUUUGAUGAUUCUCU--UACCUGCAACUCCUGUCUUUGUCGACUUUUU------ .((((....))))........((.((.(((.((((..(((-((((((((((......))))))).......--.....))))))..)))).))).)))).....------ ( -24.40) >DroPse_CAF1 52686 107 - 1 UUGGCUUUUGGCAUUAAACUUGUGCGUCAACUACACAAGU-UGCCGUCAAGUUUAGCUUUGAUGAUUCUGUUCUGCCUUCAACUCUUCU--GUCUCCACUCUUAACCUAU ..(((((..(((..((((((((.(((.(((((.....)))-)).))))))))))))))..)).((......)).)))............--................... ( -21.20) >DroEre_CAF1 30098 105 - 1 UUGCCCUUUGGCAUUAAACUUGUGCGUCAACGGCACAAGU-UGACGUCAAGUUUAGCUUUGAUGAUUCUCU--UACCUGCAACUCCUGUCUUUGGCGACUUUUU--CUGG (((((....((((.((((((((.((((((((.......))-))))))))))))))((......(.....).--.....))......))))...)))))......--.... ( -29.20) >DroYak_CAF1 32493 101 - 1 UUGCCCUUUGGCAUUAAACUUGUGCGUCAACGGCACAAGU-UGCCGUCAAGUUUAGCUUUGAUGAUUCUCU--UACCUGCAACUCCUGUCUUUGCCGACUUUUU------ .......((((((...(((((((((.......))))))))-)..(((((((......))))))).......--...................))))))......------ ( -24.60) >DroAna_CAF1 45997 101 - 1 UUGCCUUUUGGCAUUAAACUUGUGCGUCAACGACACAAGUUUGCCGUCAAGUUUAGCUUUGAUGAUUCUGU--UACCUGCAAGCCUUCUAUAU-----CUCUUU--UUGC .((((....)))).((((((((((((....)).)))))))))).(((((((......))))))).......--.....(((((..........-----.....)--)))) ( -23.76) >DroPer_CAF1 53091 107 - 1 UUGGCUUUUGGCAUUAAACUUGUGCGUCAACUACACAAGU-UGCCGUCAAGUUUAGCUUUGAUGAUUCUGUUCUGCCUUCAACUCUUCU--GUCUCCACUCUUAACCUAU ..(((((..(((..((((((((.(((.(((((.....)))-)).))))))))))))))..)).((......)).)))............--................... ( -21.20) >consensus UUGCCCUUUGGCAUUAAACUUGUGCGUCAACGACACAAGU_UGCCGUCAAGUUUAGCUUUGAUGAUUCUCU__UACCUGCAACUCCUCUCUUUGUCGACUCUUU__CU__ ((((.....((((....(((((((((....)).))))))).))))((((((......))))))...............))))............................ (-18.22 = -18.13 + -0.08)


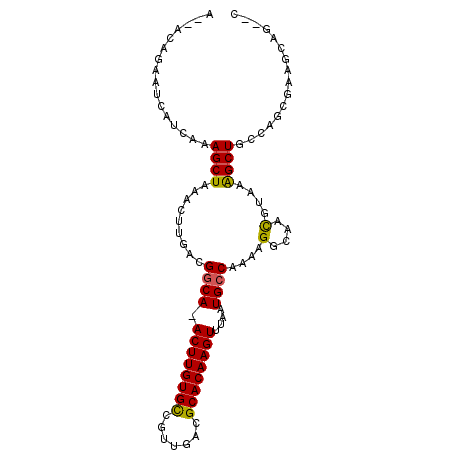
| Location | 3,898,169 – 3,898,265 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.09 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.05 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3898169 96 + 22407834 A--AGAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGCCACUGACGCACAAGUUUAAUGCCAAAGGGCAACGUAAAGCUUCUGGCGAAGCAGCCC .--.........((((........)))).((((-((((((((.......)))))))))...((((....)))).......(((((....))))).))). ( -27.80) >DroPse_CAF1 52720 92 + 1 AGAACAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGUAGUUGACGCACAAGUUUAAUGCCAAAAGCCAACGAAAAGCUGCCACCCAAA------ ....(((..((.((((........)))).((((-((((((((.......))))))))....))))..........))....))).........------ ( -17.60) >DroEre_CAF1 30132 96 + 1 A--AGAGAAUCAUCAAAGCUAAACUUGACGUCA-ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAGGGCAAUGUAAAGCUUCUGGCAAAGCAGCCC .--............((((((((((((.(((((-((.......)))))))..)))))))).((((....)))).......))))..(((......))). ( -28.30) >DroYak_CAF1 32523 96 + 1 A--AGAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAGGGCAACGUAAAGCUUCUGGCGAAGCAGUCC .--..............(((....(..((((((-.....))))))..)(((..((((((..((((....)))).....))))))...))).)))..... ( -26.40) >DroAna_CAF1 46026 96 + 1 A--ACAGAAUCAUCAAAGCUAAACUUGACGGCAAACUUGUGUCGUUGACGCACAAGUUUAAUGCCAAAAGGCAACGUAAGGCUGCCAACGAAGCCAG-G .--..............(((.........((((((((((((.((....))))))))))...))))....((((.(....)..)))).....)))...-. ( -27.70) >DroPer_CAF1 53125 92 + 1 AGAACAGAAUCAUCAAAGCUAAACUUGACGGCA-ACUUGUGUAGUUGACGCACAAGUUUAAUGCCAAAAGCCAACGAAAAGCUGCCACCCAAA------ ....(((..((.((((........)))).((((-((((((((.......))))))))....))))..........))....))).........------ ( -17.60) >consensus A__ACAGAAUCAUCAAAGCUAAACUUGACGGCA_ACUUGUGCCGUUGACGCACAAGUUUAAUGCCAAAAGGCAACGUAAAGCUGCCAGCGAAGCAG__C ................((((.........((((.((((((((.......))))))))....))))....(....)....))))................ (-18.41 = -18.05 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:27 2006