
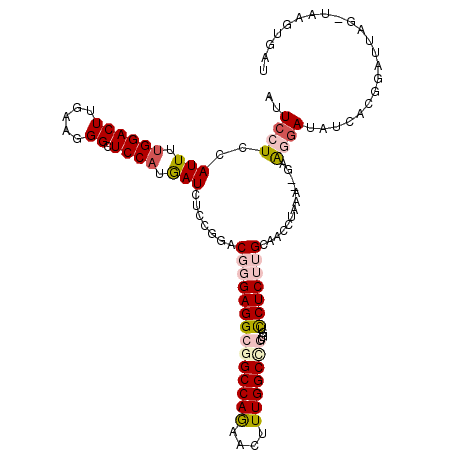
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,873,241 – 3,873,375 |
| Length | 134 |
| Max. P | 0.957596 |

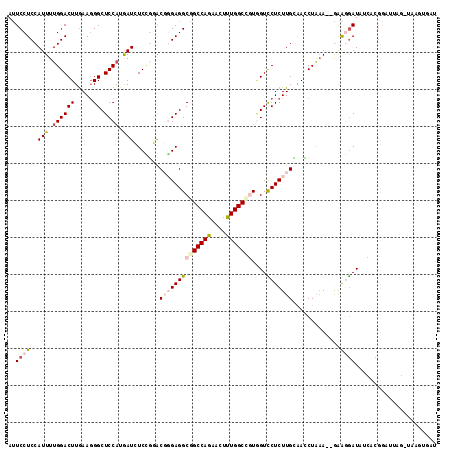
| Location | 3,873,241 – 3,873,355 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.45 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3873241 114 + 22407834 ACUCCUCCAUUUUGGACUUGAAGGGCUCCAUGAUCUCGGGACGCGAGGCGGCCAAAACUUUGGCCGUCGUUCUCUUGCAACCUGAA--GAGCAAUAUCAUGGAUUAGCUAAGUGAU ..(((........))).......(((((((((((((((((..(((((((((((((....))))))))......)))))..)))).)--))......)))))))...)))....... ( -37.80) >DroPse_CAF1 16029 107 + 1 AUUCCUCCAUUUUGGACUUGAAGGGCUCCACAAUGUCUUCACUGGAGGCAGCCAGUACCUUGGCCUUGGUCCUCUUGAAAACUAAACCAAAGGAUACGCCGAUUAAU--------- ..((((....(((((..((.(((((..(((...((((((.....))))))(((((....)))))..)))..))))).))..)))))....)))).............--------- ( -27.30) >DroEre_CAF1 5068 114 + 1 AUUCCUCCAUUUUGGACUUGAAGGGCUCCAUGAUCUCCGGGCGGGAGGCGGCCAGAACUUUGGCCGUGGUCCUCUUGCAACCUGUA--GGAUGAUUUCAUGGAUUAGCUUAGUGAU .(((.(((.....)))...)))(((((((((((..((...(((((((((((((((....)))))))....))))))))..((....--))..))..)))))))...))))...... ( -39.90) >DroYak_CAF1 7353 114 + 1 AUUCCUCCAUUUUGGACUUGAAUGGCUCCAUGAUCUCCGGACGGGAGGCGGCCAACACUUUGGCCGUAGUCCUCUUGCAACCUGAA--GGGGGAUUUCAUGGAUUAGUUAUGUGAU ......((((((.......)))))).(((((((.((((.....))))((((((((....))))))))(((((((((.(.....).)--)))))))))))))))............. ( -40.20) >DroAna_CAF1 19521 115 + 1 ACUCCUCCAUUUUGGACUUAAAGGGCUCCUGGAUAUCCGGACGCGAGGCGGCCAGCACUUUGGCUGUGGUUCUCUUGUGAUCUAAAU-GGAGGAUAUUUCUUUUUAGGAAACAGUU ..((((((((((.(((....(((((..(((((....)))..(....)((((((((....))))))))))..)))))....)))))))-))))))............(....).... ( -38.40) >DroPer_CAF1 16492 107 + 1 AUUCCUCCAUUUUGGACUUGAAGGGCUCCACAAUGUCUUCACUGGAGGCAGCCAGUACCUUGGCCUUGGUCCUCUUGAAAACUAAACCAAAGGAUACGCCGAUUAAU--------- ..((((....(((((..((.(((((..(((...((((((.....))))))(((((....)))))..)))..))))).))..)))))....)))).............--------- ( -27.30) >consensus AUUCCUCCAUUUUGGACUUGAAGGGCUCCAUGAUCUCCGGACGGGAGGCGGCCAGAACUUUGGCCGUGGUCCUCUUGCAACCUAAA__GAAGGAUAUCACGGAUUAG_UAAGUGAU ..((((..(((.((((((.....)).)))).))).......((((((((((((((....)))))))....))))))).............))))...................... (-18.42 = -19.45 + 1.03)

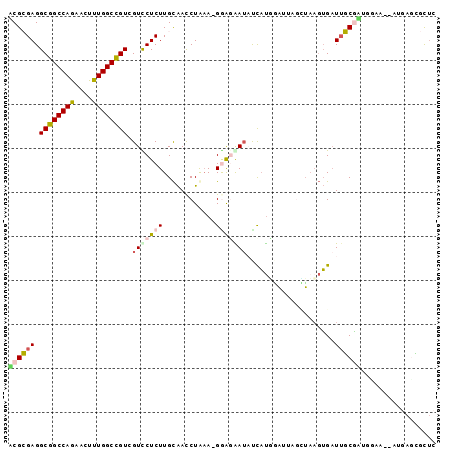
| Location | 3,873,281 – 3,873,375 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3873281 94 + 22407834 ACGCGAGGCGGCCAAAACUUUGGCCGUCGUUCUCUUGCAACCUGAA-GAGCAAUAUCAUGGAUUAGCUAAGUGAUGGCAAUGGAA--AUGAUCGCUC ..(((((((((((((....))))))))).((((.((((........-.(((...(((...)))..)))........)))).))))--....)))).. ( -31.23) >DroSec_CAF1 5671 90 + 1 UCGCGAGGCGGCCAGAACUUUGGCCGUCGUCCUCUUGCAACCUAAA-GGAGAAAACCAUUGA----UUAAGUGAUUGCGAGGGAA--AUGAGCGCUC ..(((.(((((((((....))))))))).(((.(((((((((....-)).......((((..----...)))).)))))))))).--.....))).. ( -34.60) >DroSim_CAF1 5760 90 + 1 UCGCGAGGCGGCCAGAACUUUGGCCGUCGUCCUCUUGCAACCUAAA-GAAGAAUACCAUUAA----AUAAGUGAUUGCGAAGGAU--AUGAGCGCUC ..(((.(((((((((....)))))))))(((((.((((((.((...-..)).....((((..----...)))).)))))))))))--.....))).. ( -30.10) >DroEre_CAF1 5108 94 + 1 GCGGGAGGCGGCCAGAACUUUGGCCGUGGUCCUCUUGCAACCUGUA-GGAUGAUUUCAUGGAUUAGCUUAGUGAUUUCGCUGCAA--AUUAGAGCUC (((((((((((((((....)))))))....))))))))....((((-(..(((..((((.((.....)).))))..)))))))).--.......... ( -31.30) >DroYak_CAF1 7393 94 + 1 ACGGGAGGCGGCCAACACUUUGGCCGUAGUCCUCUUGCAACCUGAA-GGGGGAUUUCAUGGAUUAGUUAUGUGAUUUCGUUGCAA--AUGAGAACUC ..(((..((((((((....))))))))..)))..((((((((....-))(..(((.(((((.....))))).)))..).))))))--.......... ( -30.80) >DroAna_CAF1 19561 97 + 1 ACGCGAGGCGGCCAGCACUUUGGCUGUGGUUCUCUUGUGAUCUAAAUGGAGGAUAUUUCUUUUUAGGAAACAGUUUGCUUAAAAAACAAUAUUGUGU ((((((.((((((((....))))))))(((....((((..((((((.((((.....)))).))))))..))))...)))............)))))) ( -23.60) >consensus ACGCGAGGCGGCCAGAACUUUGGCCGUCGUCCUCUUGCAACCUAAA_GGAGAAUAUCAUGGAUUAGCUAAGUGAUUGCGAUGGAA__AUGAGCGCUC ((((((.((((((((....)))))))).(((((((............)))))))....................))))))................. (-17.33 = -17.50 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:20 2006