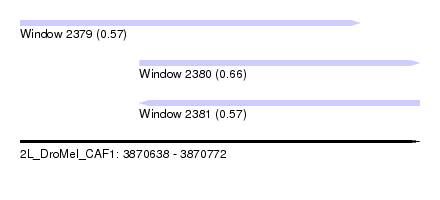
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,870,638 – 3,870,772 |
| Length | 134 |
| Max. P | 0.663273 |

| Location | 3,870,638 – 3,870,752 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -40.99 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.87 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3870638 114 + 22407834 UGCUACUUACGAAGGCCCAGCUCGAGAACGAGCGAAAGAUUCAGCAGCUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGGA---GGUGGAUCCCAAGGGAA--- ...............(((.(((((....)))))......(((.(((((.(((((((.....))))..)))))))).)))...((..((((((.....---))))))..))..)))..--- ( -41.80) >DroVir_CAF1 25091 114 + 1 UGCUGCUGACUAAGGCGCAGCUGCAGAACGAGCGCAAAAUCCAACAGCUUCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCUACCAUGCA---AGUAGAUCCCAAGGCUG--- (((.((((.(......))))).))).....(((......((((.((((....((((.....)))).....))))))))....((..(((((......---.)))))..))...))).--- ( -32.80) >DroPse_CAF1 12723 120 + 1 UGCGGCUGACCAAGGCGCAGCUGGAGAACGAGCGCAAGAUCCAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCAUGUGGUGGAUCCUAAGGCAGCAG (((.(((..(((.((((((((((......((.(....).))...)))))))....(((.........)))))).)))....(((.(((((((((....))))))))))))..))).))). ( -49.10) >DroWil_CAF1 20511 114 + 1 UGCUGCUGACCAAGGCGCAGCUGGAGAACGAACGAAAGAUUCAACAGUUGCAAAUGCAGGCCAUUAAUGCACUGUGGAAGAAGGUUUCCACAAUGGAAAUUGUGGACACAAAAG------ ..((.((..((.....(((((((..(((....(....).)))..)))))))....((((((.......)).)))))).)).))((.((((((((....)))))))).)).....------ ( -35.70) >DroMoj_CAF1 16339 114 + 1 UGAUAAUGACCAAGGUGCAGCUGCAGAACGAGCGCAAGAUACAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCA---GGUGGACCCGAAGGCUG--- ......((....(..((((((((.........(....)......))))))))..).))((((.((......((.....))..((.(((((((.....---))))))))))).)))).--- ( -37.46) >DroPer_CAF1 13315 120 + 1 UGCGGCUGACCAAGGCGCAGCUGGAGAACGAGCGCAAGAUCCAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCAUGUGGUGGAUCCUAAGGCAGCAG (((.(((..(((.((((((((((......((.(....).))...)))))))....(((.........)))))).)))....(((.(((((((((....))))))))))))..))).))). ( -49.10) >consensus UGCUGCUGACCAAGGCGCAGCUGGAGAACGAGCGCAAGAUCCAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCA___GGUGGAUCCCAAGGCAG___ .........(((.((((((((((......((.(....).))...)))))))....(((.........)))))).))).....((.(((((((........)))))))))........... (-27.33 = -27.87 + 0.53)

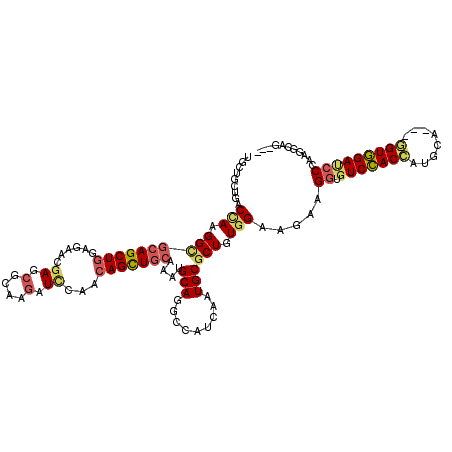
| Location | 3,870,678 – 3,870,772 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3870678 94 + 22407834 UCAGCAGCUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGGA---GGUGGAUCCCAAGGGAA---UACCUGCUGCUGCGGAACAG .(((((((.((((.((((.........))))))))......(((((((((((.....---))))))(((....))).---))))))))))))........ ( -37.70) >DroGri_CAF1 14348 88 + 1 UCAACAGCUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGCA---GGUCGAACCCAAAGCUG---AACAG----GCUGAGAAC-- ....(((((.(((.((..((((.....((((..(((((........)))))..))))---)))).....))...)))---....)----)))).....-- ( -23.50) >DroYak_CAF1 2306 94 + 1 UCAGCAGCUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGGA---GGUUGAUCCCAAGGGAA---UACCAGCUACUGGGGAACAG ((.(((((.(((((((.....))))..)))))))).)).....((.(((.((((((.---(((...(((....))).---.)))..))).)))))))).. ( -28.60) >DroMoj_CAF1 16379 85 + 1 ACAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCA---GGUGGACCCGAAGGCUG------------GCGGCGAGCAC ......(((((....((.((((.((......((.....))..((.(((((((.....---))))))))))).)))).------------)).)).))).. ( -32.00) >DroAna_CAF1 17177 94 + 1 CCAGCAACUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGCA---AGUGGACCCCAAAGGCG---UGCCUCCAGCCGUGGACCAG .((((..((((....))))((.......))))))(((.....((.((((((......---.)))))).))...(((.---........))).....))). ( -30.80) >DroPer_CAF1 13355 100 + 1 CCAACAGCUGCAAAUGCAGGCCAUCAAUGCGCUGUGGAAGAAGGUGUCCACCAUGCAUGUGGUGGAUCCUAAGGCAGCAGGACAAGUCGCCGCAGUUUCG .....((((((......((((((((((((((..(((((........)))))..))))).))))))..)))..(((.((.......)).)))))))))... ( -35.10) >consensus UCAACAGCUGCAGAUGCAGGCCAUCAAUGCGCUGUGGAAAAAGGUGUCCACCAUGCA___GGUGGAUCCCAAGGCAG___UACCAGC_GCCGGGGAACAG ...(((((.(((((((.....))))..)))))))).......((.(((((((........))))))).)).............................. (-19.86 = -20.67 + 0.81)

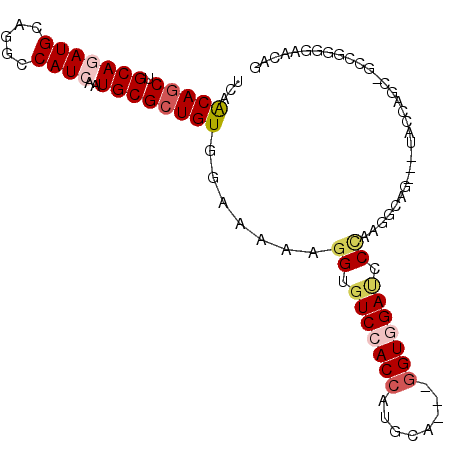
| Location | 3,870,678 – 3,870,772 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3870678 94 - 22407834 CUGUUCCGCAGCAGCAGGUA---UUCCCUUGGGAUCCACC---UCCAUGGUGGACACCUUUUUCCACAGCGCAUUGAUGGCCUGCAUCUGCAGCUGCUGA ........(((((((((((.---.(((....)))((((((---.....)))))).))))...........(((..((((.....))))))).))))))). ( -36.10) >DroGri_CAF1 14348 88 - 1 --GUUCUCAGC----CUGUU---CAGCUUUGGGUUCGACC---UGCAUGGUGGACACCUUUUUCCACAGCGCAUUGAUGGCCUGCAUCUGCAGCUGUUGA --.......((----((((.---.......((((((.(((---.....))).)).))))......)))).)).(..(((((.(((....))))))))..) ( -25.94) >DroYak_CAF1 2306 94 - 1 CUGUUCCCCAGUAGCUGGUA---UUCCCUUGGGAUCAACC---UCCAUGGUGGACACCUUUUUCCACAGCGCAUUGAUGGCCUGCAUCUGCAGCUGCUGA ........(((((((((((.---.(((....)))...)))---......(((((........)))))...(((..((((.....))))))))))))))). ( -30.80) >DroMoj_CAF1 16379 85 - 1 GUGCUCGCCGC------------CAGCCUUCGGGUCCACC---UGCAUGGUGGACACCUUCUUCCACAGCGCAUUGAUGGCCUGCAUUUGCAGCUGUUGU (((((.((...------------..))....(((((((((---.....))))))).)).........)))))...((((((.(((....))))))))).. ( -30.80) >DroAna_CAF1 17177 94 - 1 CUGGUCCACGGCUGGAGGCA---CGCCUUUGGGGUCCACU---UGCAUGGUGGACACCUUUUUCCACAGCGCAUUGAUGGCCUGCAUCUGCAGUUGCUGG .(((.....(((..((((..---..))))..).(((((((---.....))))))).)).....)))((((((.......))((((....))))..)))). ( -34.20) >DroPer_CAF1 13355 100 - 1 CGAAACUGCGGCGACUUGUCCUGCUGCCUUAGGAUCCACCACAUGCAUGGUGGACACCUUCUUCCACAGCGCAUUGAUGGCCUGCAUUUGCAGCUGUUGG .(((...((((((........))))))....((.(((((((......)))))))..)))))..((((((((((..((((.....))))))).)))).))) ( -32.00) >consensus CUGUUCCCCGGC_GCAGGUA___CUCCCUUGGGAUCCACC___UGCAUGGUGGACACCUUUUUCCACAGCGCAUUGAUGGCCUGCAUCUGCAGCUGCUGA ..............................((..((((((........))))))..))......(((((((((..((((.....))))))).)))).)). (-18.67 = -18.83 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:17 2006