| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 433,927 – 434,044 |
| Length | 117 |
| Max. P | 0.635831 |

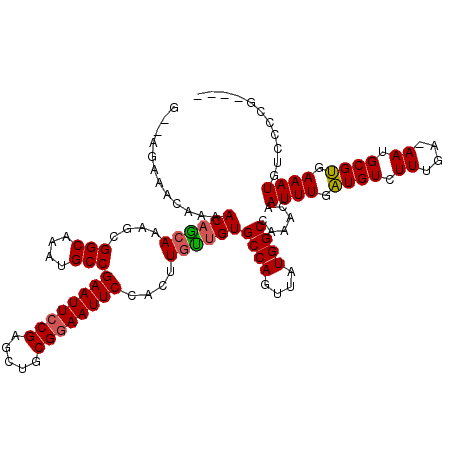
| Location | 433,927 – 434,044 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 433927 117 + 22407834 UCGGCGGGGACAUUUCACGCAUU-UCAAAGACAUCAAAUUGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUAUUUCU--C .((((((((.(......(((...-....(((((......))))).((((....)))).......)))((((((((.....))))))))(((....))))))))))))..........--. ( -34.20) >DroSec_CAF1 11332 117 + 1 UCGGCGGGGACAUUUCACGCAUU-UCAAAGACAUCAAAUUGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUUU--C .((((((((.(......(((...-....(((((......))))).((((....)))).......)))((((((((.....))))))))(((....))))))))))))..........--. ( -34.20) >DroSim_CAF1 10388 117 + 1 UCGGCGGGGACAUUUCACGCAUU-UCAAAGACAUCAAAUUGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU--C .((((((((.(......(((...-....(((((......))))).((((....)))).......)))((((((((.....))))))))(((....))))))))))))..........--. ( -34.20) >DroEre_CAF1 11438 113 + 1 ----CGGGGACAUUUCACGCAUU-UCAAAGACAUCAAAUUGUUUGGCCAUAACUGGCACAGCAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUGUGCUGUUUUGUUUCU--C ----.((..(((...((.((...-....(((((......))))).((((....))))(((((.....((((((((.....))))))))(((....))))))))))))...)))..))--. ( -39.20) >DroYak_CAF1 11026 113 + 1 ----CGGGGACAUUUCACGCAUU-UCAAAGACAUCAAAUUGUUUAGCCAUAACUGGCACAGCAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU--C ----.((..(((...........-....(((((......))))).((((....))))(((((((((.((((((((.....))))))))(((....))))).)))))))..)))..))--. ( -37.20) >DroAna_CAF1 14320 112 + 1 --------GACAUUUCGCGCAUUUUCAAAGACACCAAAUUGUUUGGCCAUAACUGGCACAACAAGUGGAAUUUCGGAAUUCGGAAUUCGGCAUUUGCCGCUGUUUGGUUUUGUUUCUGUC --------..........(((....((((...(((((((((((..((((....))))..))))(((.((((((((.....))))))))(((....)))))))))))))))))....))). ( -28.80) >consensus ____CGGGGACAUUUCACGCAUU_UCAAAGACAUCAAAUUGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU__C .....(((((((...((.(((..........(((....(((((..((((....))))..))))))))((((((((.....))))))))(((....)))....)))))...)))))))... (-28.60 = -28.85 + 0.25)

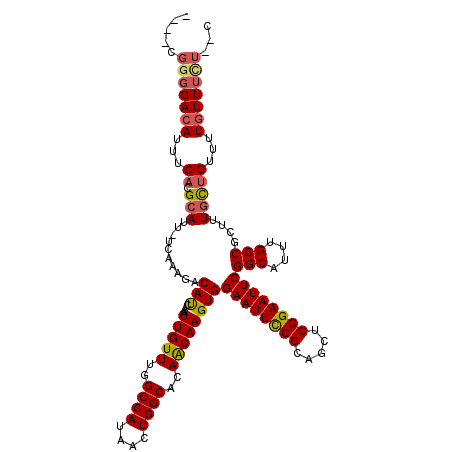
| Location | 433,927 – 434,044 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -27.90 |
| Energy contribution | -27.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 433927 117 - 22407834 G--AGAAAUAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACAAUUUGAUGUCUUUGA-AAUGCGUGAAAUGUCCCCGCCGA (--(((..(((((((((((...(((....)))((((((((.....))))))))...)))))))((((....)))).......))))...))))...-...(((.(........))))... ( -30.50) >DroSec_CAF1 11332 117 - 1 G--AAAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACAAUUUGAUGUCUUUGA-AAUGCGUGAAAUGUCCCCGCCGA .--...(((...(((((((...(((....)))((((((((.....))))))))...)))))))....))).((((......((((.((((......-...)))).))))......)))). ( -31.20) >DroSim_CAF1 10388 117 - 1 G--AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACAAUUUGAUGUCUUUGA-AAUGCGUGAAAUGUCCCCGCCGA (--(((..(((((((((((...(((....)))((((((((.....))))))))...)))))))((((....)))).......))))...))))...-...(((.(........))))... ( -32.30) >DroEre_CAF1 11438 113 - 1 G--AGAAACAAAACAGCACAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGCUGUGCCAGUUAUGGCCAAACAAUUUGAUGUCUUUGA-AAUGCGUGAAAUGUCCCCG---- .--.....((.(((.((((((((((....)))((((((((.....)))))))).....)))))))..))).))........((((.((((......-...)))).)))).......---- ( -33.40) >DroYak_CAF1 11026 113 - 1 G--AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGCUGUGCCAGUUAUGGCUAAACAAUUUGAUGUCUUUGA-AAUGCGUGAAAUGUCCCCG---- (--(((..(((((((((((...(((....)))((((((((.....))))))))...)))))))((((....)))).......))))...))))...-...................---- ( -33.10) >DroAna_CAF1 14320 112 - 1 GACAGAAACAAAACCAAACAGCGGCAAAUGCCGAAUUCCGAAUUCCGAAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACAAUUUGGUGUCUUUGAAAAUGCGCGAAAUGUC-------- ((((........(((((((((((((....)))(((((.((.....)).))))).....)))))((((....))))........)))))...((((........)))).))))-------- ( -25.90) >consensus G__AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACAAUUUGAUGUCUUUGA_AAUGCGUGAAAUGUCCCCG____ ............((((((....(((....)))((((((((.....))))))))....))))))((((....))))......((((.((((.((....)).)))).))))........... (-27.90 = -27.77 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:25 2006