| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,861,213 – 3,861,314 |
| Length | 101 |
| Max. P | 0.915417 |

| Location | 3,861,213 – 3,861,314 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3861213 101 + 22407834 -------------U----CCUCUCUGAUUUGCACAAAGUAUUUGUUUUCUUUCCGUGUAAGCUACUUGCUGUGGGCUCUUUGGAAAUGGCCCACACAAGUGCA--UUUGGGCCAUAUCUG -------------.----.......(((..((.((((((((((((..............(((.....)))(((((((.((....)).))))))))))))))).--)))).))...))).. ( -28.00) >DroSec_CAF1 49681 84 + 1 -------------U----G----CCGAUUUGCACAAAGUAUUUAUUUUCUUUCCGUGUAAGCUACU-------------UUGGAAAUGGCCCACACAAGUGCA--UUUGGGCCAUAUCUG -------------(----(----(......)))((((((((((((...........))))).))))-------------)))((.((((((((((....))..--..)))))))).)).. ( -22.60) >DroSim_CAF1 48840 84 + 1 -------------U----G----CCGGUUUGCACAAAGUAUUUAUUUUCUUUCCGUGUAAGCUACU-------------UUGGAAAUGGCCCACACAAGUGCA--UUUGGGCCAUAUCUG -------------.----.----..(((((((((((((..........))))..)))))))))...-------------..(((.((((((((((....))..--..)))))))).))). ( -25.70) >DroEre_CAF1 54781 97 + 1 -------------U----C----CUGAUUUGCACAAACCACUGGUUUUCUUUCCGUGUAAGCUACUUGUUUAGGGCUCCUUGGAAAUGGCCCACACAAGUGCA--UUUGGGCCAUAUCUG -------------.----(----(..(..((((((((((...))))).......((((((((.....)))).(((((...(....).)))))))))..)))))--.)..))......... ( -26.30) >DroYak_CAF1 52382 97 + 1 -------------U----C----CUGAUUUGCACAAAAUAUUGGUUUUCUUUCCGUGUAAGCCACUGGGUUAGGGCUCUUUGGAAAUGGCCCACACAAGUGCA--UUUGGGCCAUAUCUU -------------.----(----(..(..(((((........((........))((((.((((....)))).(((((.((....)).)))))))))..)))))--.)..))......... ( -27.60) >DroAna_CAF1 50458 114 + 1 UCUAGUUGCGAACUUCGGG----UCUAUUAGCCAGAAGUAUUUGUUUUCUUUCCGUGUAAGCUACUUGUUUAGGGC-CUUCGGAAAAGGCCCACACAGGCGCAGAUUCGAGCC-UAUCUU .............((((((----(((....((((((((.......)))))....((((((((.....)))).((((-(((.....))))))))))).)))..)))))))))..-...... ( -33.90) >consensus _____________U____C____CCGAUUUGCACAAAGUAUUUGUUUUCUUUCCGUGUAAGCUACUUG_UUAGGGC_CUUUGGAAAUGGCCCACACAAGUGCA__UUUGGGCCAUAUCUG ...........................(((((((((((..........))))..)))))))....................(((.((((((((..............)))))))).))). (-14.68 = -15.87 + 1.20)


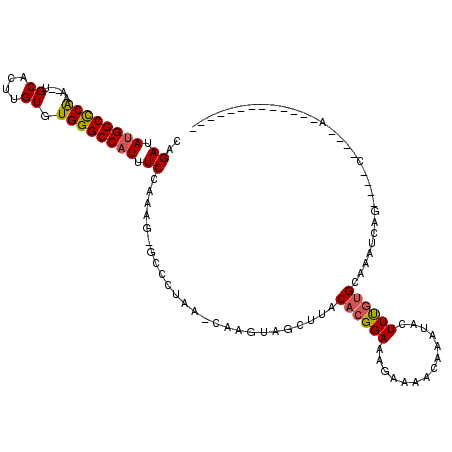
| Location | 3,861,213 – 3,861,314 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3861213 101 - 22407834 CAGAUAUGGCCCAAA--UGCACUUGUGUGGGCCAUUUCCAAAGAGCCCACAGCAAGUAGCUUACACGGAAAGAAAACAAAUACUUUGUGCAAAUCAGAGAGG----A------------- ..........((...--.((((((((((((((..((....))..))))))).))))).))......................(((((.......))))).))----.------------- ( -25.20) >DroSec_CAF1 49681 84 - 1 CAGAUAUGGCCCAAA--UGCACUUGUGUGGGCCAUUUCCAA-------------AGUAGCUUACACGGAAAGAAAAUAAAUACUUUGUGCAAAUCGG----C----A------------- ........(((....--(((((((((((((((.(((.....-------------))).))))))))))((((..........)))))))))....))----)----.------------- ( -22.30) >DroSim_CAF1 48840 84 - 1 CAGAUAUGGCCCAAA--UGCACUUGUGUGGGCCAUUUCCAA-------------AGUAGCUUACACGGAAAGAAAAUAAAUACUUUGUGCAAACCGG----C----A------------- ........(((....--(((((((((((((((.(((.....-------------))).))))))))))((((..........)))))))))....))----)----.------------- ( -22.30) >DroEre_CAF1 54781 97 - 1 CAGAUAUGGCCCAAA--UGCACUUGUGUGGGCCAUUUCCAAGGAGCCCUAAACAAGUAGCUUACACGGAAAGAAAACCAGUGGUUUGUGCAAAUCAG----G----A------------- ..(((...((.((((--(((((((((..((((....((....))))))...)))))).))...(((((........)).)))))))).))..)))..----.----.------------- ( -28.00) >DroYak_CAF1 52382 97 - 1 AAGAUAUGGCCCAAA--UGCACUUGUGUGGGCCAUUUCCAAAGAGCCCUAACCCAGUGGCUUACACGGAAAGAAAACCAAUAUUUUGUGCAAAUCAG----G----A------------- .....((((((((..--.((....)).)))))))).(((...(((((((.....)).))))).(((((((............))))))).......)----)----)------------- ( -26.60) >DroAna_CAF1 50458 114 - 1 AAGAUA-GGCUCGAAUCUGCGCCUGUGUGGGCCUUUUCCGAAG-GCCCUAAACAAGUAGCUUACACGGAAAGAAAACAAAUACUUCUGGCUAAUAGA----CCCGAAGUUCGCAACUAGA .(.(((-(((.((....)).)))))).)((((((((...))))-))))......(((.((.....(.....).........(((((.(((.....).----)).)))))..)).)))... ( -29.20) >consensus CAGAUAUGGCCCAAA__UGCACUUGUGUGGGCCAUUUCCAAAG_GCCCUAA_CAAGUAGCUUACACGGAAAGAAAACAAAUACUUUGUGCAAAUCAG____C____A_____________ ..((.((((((((.....((....)).)))))))).)).........................((((((..............))))))............................... (-14.74 = -14.82 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:09 2006