
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,858,068 – 3,858,199 |
| Length | 131 |
| Max. P | 0.978707 |

| Location | 3,858,068 – 3,858,168 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -30.68 |
| Energy contribution | -31.07 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
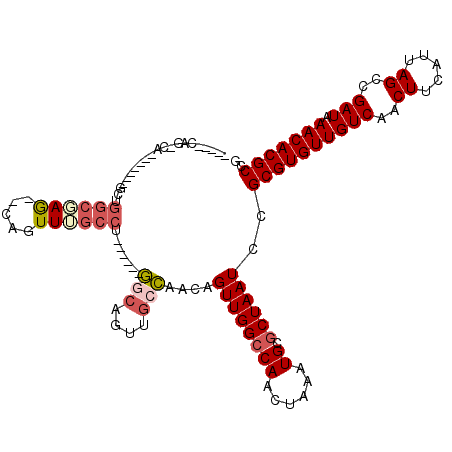
>2L_DroMel_CAF1 3858068 100 + 22407834 UCCUCCU--ACU-------------GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC----- .......--...-------------(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).)))))))........(((....)))----- ( -34.40) >DroSim_CAF1 45779 100 + 1 UCCUCCU--ACU-------------GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC----- .......--...-------------(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).)))))))........(((....)))----- ( -34.40) >DroEre_CAF1 51657 100 + 1 UCCUCCU--ACU-------------GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGGC----- ....((.--...-------------(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).)))))))........(....).)).----- ( -32.40) >DroWil_CAF1 85561 117 + 1 UCCCUCG--ACUGCAACCAGAGUCCGCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUG-CCAACUGGCCUCGACUUCGGCUCU ......(--(((........)))).(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).))))(-((....))).........)))... ( -34.60) >DroYak_CAF1 49167 100 + 1 UCCUCCU--ACU-------------GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC----- .......--...-------------(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).)))))))........(((....)))----- ( -34.40) >DroAna_CAF1 47313 102 + 1 UCUCCCUGCGCU-------------GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC----- ......((((((-------------(............(((.(((((((..((((((.....)))))).))))))).))))))))))..((((((.(((.....)))))))))..----- ( -34.90) >consensus UCCUCCU__ACU_____________GCCAAUUUAAUGUGUCGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC_____ .........................(((((((.((((((((.(((((((..((((((.....)))))).))))))).)....))))))).)))))))........(((....)))..... (-30.68 = -31.07 + 0.39)


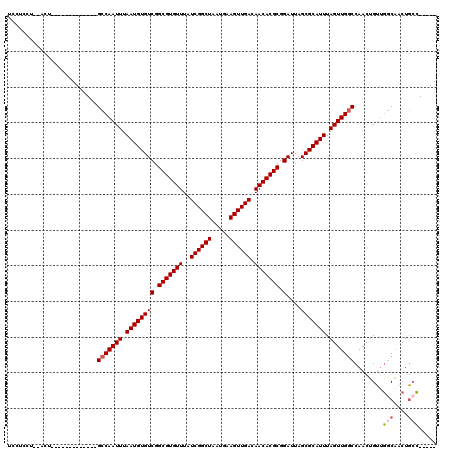
| Location | 3,858,093 – 3,858,199 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -23.13 |
| Energy contribution | -24.44 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3858093 106 + 22407834 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC------AGGCAAACGG--UUCGCCAGCAGAGUGCUG-CUG----- (.(((((((..((((((.....)))))).))))))).)..((((((((((.((((((.((((((((((....)))------).....))))--)).)))))).)))))).)-)))----- ( -47.60) >DroSec_CAF1 46705 106 + 1 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC------AGGCAAACUG--CUGGCCAGCAUAGUGCUG-CUG----- (.(((((((..((((((.....)))))).))))))).)..(((((((((..((((((((....(((((....)))------))((.....)--)))))))))..))))).)-)))----- ( -45.60) >DroEre_CAF1 51682 94 + 1 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGGC------AGGCAAACUG--CUCGCCAGC------------------ (.(((((((..((((((.....)))))).))))))).)......((...(((((....)))))...))..(((((------..((.....)--)..))))).------------------ ( -34.30) >DroWil_CAF1 85599 99 + 1 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUG-CCAACUGGCCUCGACUUCGGCUCUGCGGCCAACUGGCCUGGCCA-------------------- .((((((((..((((((.....)))))).)))))(((......)))........)-))....((((..(((....).))...((((....)))).)))).-------------------- ( -36.30) >DroYak_CAF1 49192 112 + 1 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC------AGGCAAACUG--UUCGCCAGCAGAGUGCUGGCUGGCAUA (.(((((((..((((((.....)))))).))))))).).(((((((((((.((((((.((((((((((....)))------).)))....)--)).)))))).))))))..))))).... ( -43.90) >DroAna_CAF1 47340 105 + 1 CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC------GAGGCAACUG--CUUGGGGAC-------UGGGCGGAUUG ..(((((((..((((((.....)))))).))))))).((((.((.((..((((((.(((.....)))))))))((------.((((....)--))).))...-------)).)).)))). ( -37.00) >consensus CGGCGUGUUUAUCGGCUAAUGAAGUUGACAACACGCGGAUUAGCGCAUUUAGUUGGCCAACUGUUGGCAACUGCC______AGGCAAACUG__CUCGCCAGC_______UG_CUG_____ (.(((((((..((((((.....)))))).))))))).)....(((....(((((.(((.......(((....))).......))).)))))....)))...................... (-23.13 = -24.44 + 1.31)

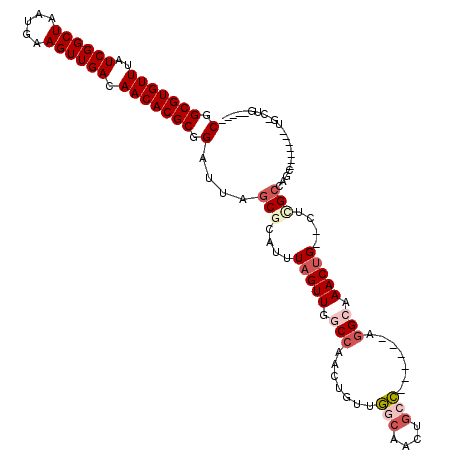
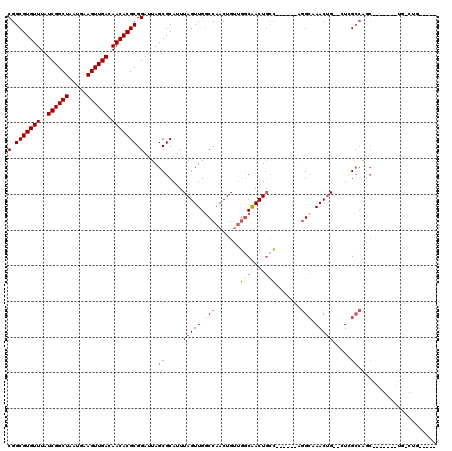
| Location | 3,858,093 – 3,858,199 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -20.38 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
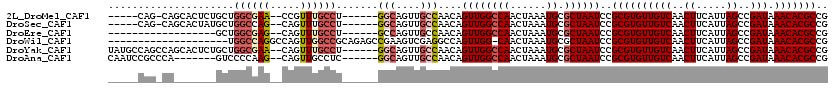
>2L_DroMel_CAF1 3858093 106 - 22407834 -----CAG-CAGCACUCUGCUGGCGAA--CCGUUUGCCU------GGCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG -----.((-(((....)))))((((..--..(((((..(------(((((((((((.....))).))))).(((((((......))))))).............)))).))))).)))). ( -34.10) >DroSec_CAF1 46705 106 - 1 -----CAG-CAGCACUAUGCUGGCCAG--CAGUUUGCCU------GGCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG -----.((-(.(((....(.(((((((--(.((.....(------(((....)))))).)))))))).)....)))))).....((((((((((..((.....))..))).))))))).. ( -36.20) >DroEre_CAF1 51682 94 - 1 ------------------GCUGGCGAG--CAGUUUGCCU------GCCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG ------------------(((((((.(--(.....)).)------))))))........((((((((......)).))))))..((((((((((..((.....))..))).))))))).. ( -29.20) >DroWil_CAF1 85599 99 - 1 --------------------UGGCCAGGCCAGUUGGCCGCAGAGCCGAAGUCGAGGCCAGUUGG-CAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG --------------------.(((..((((....))))((.(.(((........))))((((((-((((...(((((......)))))))))))))))......))..........))). ( -36.10) >DroYak_CAF1 49192 112 - 1 UAUGCCAGCCAGCACUCUGCUGGCGAA--CAGUUUGCCU------GGCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG ...((((((((((.....))))))...--.((((.((((------(((....))))......))).))))...)).))......((((((((((..((.....))..))).))))))).. ( -37.10) >DroAna_CAF1 47340 105 - 1 CAAUCCGCCCA-------GUCCCCAAG--CAGUUGCCUC------GGCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG ...........-------........(--(.(((...((------(((((((((((.....))).))))).(((((((......))))))).............)))))..)))..)).. ( -25.80) >consensus _____CAG_CA_______GCUGGCGAG__CAGUUUGCCU______GGCAGUUGCCAACAGUUGGCCAACUAAAUGCGCUAAUCCGCGUGUUGUCAACUUCAUUAGCCGAUAAACACGCCG .....................((((((.....)))))).......(((....)))....((((((((......)).))))))..((((((((((..((.....))..))).))))))).. (-20.38 = -21.22 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:07 2006