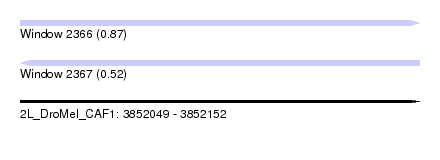
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,852,049 – 3,852,152 |
| Length | 103 |
| Max. P | 0.865505 |

| Location | 3,852,049 – 3,852,152 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -37.21 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
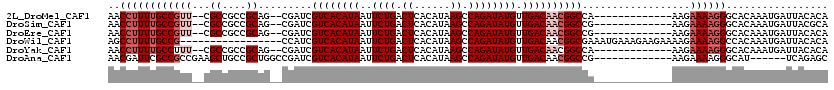
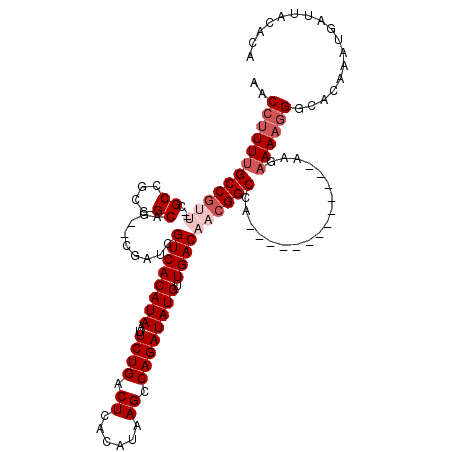
>2L_DroMel_CAF1 3852049 103 + 22407834 UGUGUAAUCAUUUGUGCCCUUUUCUU-------------UGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCG--CUGCGGCGGCG--AACGGCAAAAGGUU .(..(((....)))..).....((((-------------(.((((((((((.((((((((((((....))))))))..))))))))))(((--(((...)))))--).)))).))))).. ( -36.20) >DroSim_CAF1 40003 103 + 1 UGCGUAAUCAUUUGUGCCCUUUUCUU-------------CGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCG--CUGCGGCGGCG--AACGGCAAAAGGUU .((((((....))))))((((((...-------------..((((((((((.((((((((((((....))))))))..))))))))))(((--(((...)))))--).)))))))))).. ( -36.10) >DroEre_CAF1 45893 103 + 1 UGUGUAAUCAUUUGUGCCCUUUUCUU-------------CGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCG--CUGCGGCGGCG--AACGGCAAAAGGUU .(..(((....)))..)((((((...-------------..((((((((((.((((((((((((....))))))))..))))))))))(((--(((...)))))--).)))))))))).. ( -36.10) >DroWil_CAF1 74958 103 + 1 UGUGUAAUCAUUUGUGGCCUUUUCUUUUCUUCUUUCAUUUCGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUGG-----------------CGGCAAAAGGCU ...............((((((((................((((((((((((.((((((((((((....))))))))..)))))))))))))-----------------))).)))))))) ( -36.63) >DroYak_CAF1 42823 103 + 1 UGUGUAAUCAUUUGUGCCCUUUUCUU-------------UGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCG--CUGCGGCGGCG--AAAGGCAAAAGGUU .(..(((....)))..)(((((((((-------------(.((((((((((.((((((((((((....))))))))..))))))))))).(--(....))))).--)))))..))))).. ( -36.80) >DroAna_CAF1 41346 101 + 1 GCUCUGA------AUGCCCUUUUCUU-------------CGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCGGCCAGCGGCAGCUUCGGCGGCGAAUCGUU (((((((------(((((((......-------------.(((((((((((.((((((((((((....))))))))..)))))))))).))))))).))))..))))).)))........ ( -41.41) >consensus UGUGUAAUCAUUUGUGCCCUUUUCUU_____________CGGCCGUUGUCAACAUAUCUGGCUUAUGUGAGUCAGAAUUAUGUGACGAUCG__CUGCGGCGGCG__AACGGCAAAAGGUU .........................................((((((((((.((((((((((((....))))))))..)))))))))))......((....))......)))........ (-24.50 = -24.58 + 0.08)

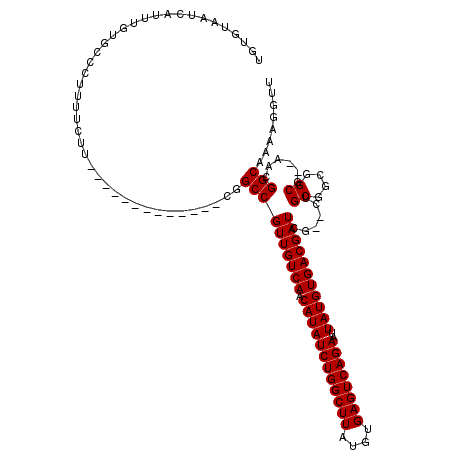
| Location | 3,852,049 – 3,852,152 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -16.99 |
| Energy contribution | -18.41 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3852049 103 - 22407834 AACCUUUUGCCGUU--CGCCGCCGCAG--CGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCA-------------AAGAAAAGGGCACAAAUGAUUACACA ..((((((((((((--(((.......)--)))..((((((((..((((.((......)).)))))))).)))).)))))..-------------...))))))................. ( -26.50) >DroSim_CAF1 40003 103 - 1 AACCUUUUGCCGUU--CGCCGCCGCAG--CGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCG-------------AAGAAAAGGGCACAAAUGAUUACGCA ..((((((((((((--(((.......)--)))..((((((((..((((.((......)).)))))))).)))).)))))..-------------...))))))................. ( -26.50) >DroEre_CAF1 45893 103 - 1 AACCUUUUGCCGUU--CGCCGCCGCAG--CGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCG-------------AAGAAAAGGGCACAAAUGAUUACACA ..((((((((((((--(((.......)--)))..((((((((..((((.((......)).)))))))).)))).)))))..-------------...))))))................. ( -26.50) >DroWil_CAF1 74958 103 - 1 AGCCUUUUGCCG-----------------CCAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCGAAAUGAAAGAAGAAAAGAAAAGGCCACAAAUGAUUACACA .(((((((..((-----------------((...((((((((..((((.((......)).)))))))).))))...))))...(....)........)))))))................ ( -23.70) >DroYak_CAF1 42823 103 - 1 AACCUUUUGCCUUU--CGCCGCCGCAG--CGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCA-------------AAGAAAAGGGCACAAAUGAUUACACA ..((((((..((((--.(((((....)--)....((((((((..((((.((......)).)))))))).))))...))).)-------------))).))))))................ ( -25.90) >DroAna_CAF1 41346 101 - 1 AACGAUUCGCCGCCGAAGCUGCCGCUGGCCGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCG-------------AAGAAAAGGGCAU------UCAGAGC ...((...(((.....(((....)))(((((...((((((((..((((.((......)).)))))))).))))..))))).-------------........)))..------))..... ( -28.40) >consensus AACCUUUUGCCGUU__CGCCGCCGCAG__CGAUCGUCACAUAAUUCUGACUCACAUAAGCCAGAUAUGUUGACAACGGCCA_____________AAGAAAAGGGCACAAAUGAUUACACA ..((((((((((((...((....)).........((((((((..((((.((......)).)))))))).))))))))))..................))))))................. (-16.99 = -18.41 + 1.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:04 2006