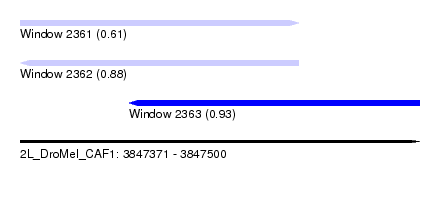
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,847,371 – 3,847,500 |
| Length | 129 |
| Max. P | 0.931024 |

| Location | 3,847,371 – 3,847,461 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -17.48 |
| Consensus MFE | -5.51 |
| Energy contribution | -5.57 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3847371 90 + 22407834 CAAAAAGUCA-CAAACAAA-UGCGGGAAAAAAACCUUUGAACAAAA-AAAAACGACUUG-GCU-----------AAAAAGCGGGUUAA-----------AACCGCUUCUCGUAAAU ..........-........-((((((((.......((((..(((..-.........)))-..)-----------)))..((((.....-----------..))))))))))))... ( -16.30) >DroSec_CAF1 36118 91 + 1 CAAAAAGUCA-CAAACAAA-UGCGGGUAAAAAACCUUUGAAAAAAACAAAAACGACUUG-GCU-----------AAAAAGCGGCUUAA-----------AACCGCUUCUCGUAAAU ....(((((.-........-....(((.....)))(((....)))........))))).-...-----------...((((((.....-----------..))))))......... ( -13.80) >DroSim_CAF1 35419 88 + 1 CAAAAAGUCA-CAAACAAA-UGCGGGUAAAAAACCUUUGAAAAA----AAAACGACUUGGGCU-----------AAAAAGCGGCUUAA-----------AACCGCUUCUCGUAAAU ....(((((.-........-....(((.....)))(((....))----)....)))))(((..-----------...((((((.....-----------..)))))))))...... ( -14.70) >DroEre_CAF1 41047 96 + 1 CAAAGAGGCA-CAAAUAAAUUGCGGGAAAAAAACCUUUGAGUAAGA-------GACUCG-GCUAAAAAGCGACUAAAAAGCGGCUUAA-----------AACCGCUUCUCGUAAAU ..........-........(((((((((.........(((((....-------.)))))-(((....))).........((((.....-----------..))))))))))))).. ( -21.10) >DroYak_CAF1 38046 86 + 1 CAAAAAGUCA-CAAAUAAA-UGCGGUUAAAAAACCUUUGAAAAAGA-------GACUUG-GCUUAAAA---------ACGCGGCCUAA-----------AACCGCUUCUCGUAAAU ..........-........-.((((((.......((((.....)))-------)....(-(((.....---------....))))...-----------))))))........... ( -14.20) >DroAna_CAF1 37238 90 + 1 CGGAAAGGCAGCCAGCAAA-CGAGGGAAAAAA-UCUACA---------AAAAG---GUG-GCU-----------AAAAAGCGGCUUAGAGCCGGGCUCAGACUGCCUCUCGUAAAU ..((.((((((((..(...-.)..))......-(((...---------...((---((.-(((-----------....))).)))).((((...))))))))))))).))...... ( -24.80) >consensus CAAAAAGUCA_CAAACAAA_UGCGGGAAAAAAACCUUUGAAAAA_A__AAAACGACUUG_GCU___________AAAAAGCGGCUUAA___________AACCGCUUCUCGUAAAU .......................((........))..........................................((((((..................))))))......... ( -5.51 = -5.57 + 0.06)

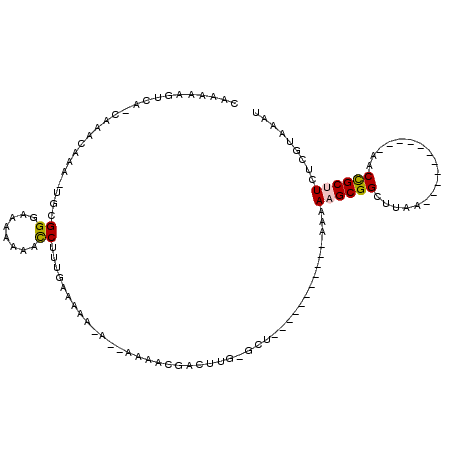
| Location | 3,847,371 – 3,847,461 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -7.79 |
| Energy contribution | -8.74 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3847371 90 - 22407834 AUUUACGAGAAGCGGUU-----------UUAACCCGCUUUUU-----------AGC-CAAGUCGUUUUU-UUUUGUUCAAAGGUUUUUUUCCCGCA-UUUGUUUG-UGACUUUUUG ......(((((((((..-----------.....)))))))))-----------...-.(((((......-...........((........))(((-......))-)))))).... ( -16.90) >DroSec_CAF1 36118 91 - 1 AUUUACGAGAAGCGGUU-----------UUAAGCCGCUUUUU-----------AGC-CAAGUCGUUUUUGUUUUUUUCAAAGGUUUUUUACCCGCA-UUUGUUUG-UGACUUUUUG ......((((((((((.-----------....))))))))))-----------...-.(((((..................(((.....))).(((-......))-)))))).... ( -20.40) >DroSim_CAF1 35419 88 - 1 AUUUACGAGAAGCGGUU-----------UUAAGCCGCUUUUU-----------AGCCCAAGUCGUUUU----UUUUUCAAAGGUUUUUUACCCGCA-UUUGUUUG-UGACUUUUUG ......((((((((((.-----------....))))))))))-----------.....(((((.....----.........(((.....))).(((-......))-)))))).... ( -20.40) >DroEre_CAF1 41047 96 - 1 AUUUACGAGAAGCGGUU-----------UUAAGCCGCUUUUUAGUCGCUUUUUAGC-CGAGUC-------UCUUACUCAAAGGUUUUUUUCCCGCAAUUUAUUUG-UGCCUCUUUG ......((((..(((((-----------..((((.(((....))).))))...)))-))..))-------)).....((((((.........(((((.....)))-))..)))))) ( -24.20) >DroYak_CAF1 38046 86 - 1 AUUUACGAGAAGCGGUU-----------UUAGGCCGCGU---------UUUUAAGC-CAAGUC-------UCUUUUUCAAAGGUUUUUUAACCGCA-UUUAUUUG-UGACUUUUUG ......((((((((((.-----------....))))).)---------))))....-.(((((-------...........((((....))))(((-......))-)))))).... ( -17.90) >DroAna_CAF1 37238 90 - 1 AUUUACGAGAGGCAGUCUGAGCCCGGCUCUAAGCCGCUUUUU-----------AGC-CAC---CUUUU---------UGUAGA-UUUUUUCCCUCG-UUUGCUGGCUGCCUUUCCG ......(((((((((((..(((..(((.....)))(((....-----------)))-...---.....---------......-............-...)))))))))))))).. ( -27.40) >consensus AUUUACGAGAAGCGGUU___________UUAAGCCGCUUUUU___________AGC_CAAGUCGUUUU__U_UUUUUCAAAGGUUUUUUACCCGCA_UUUGUUUG_UGACUUUUUG ......(((((((((((..............))))))))))).......................................................................... ( -7.79 = -8.74 + 0.95)



| Location | 3,847,406 – 3,847,500 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3847406 94 - 22407834 CU-CCCAACCUCUCCCCUAUUCCCCCUUUCCACGCUUGCGAUUUACGAGAAGCGGUUUUAACCCGCUUUUUAGC-CAAGUCGUUUUU-UUUUGUUCA ..-..................................(((((((..(((((((((.......)))))))))...-.)))))))....-......... ( -16.00) >DroSec_CAF1 36153 96 - 1 CAACCCGACCACUCCCCUACACCCCUUUUCCACGCUUGCGAUUUACGAGAAGCGGUUUUAAGCCGCUUUUUAGC-CAAGUCGUUUUUGUUUUUUUCA ...............................(((...(((((((..((((((((((.....))))))))))...-.)))))))...)))........ ( -19.40) >DroSim_CAF1 35454 93 - 1 CAACCCAGCCACUCCCCUACACCCCUUUUCCACGCUUGCGAUUUACGAGAAGCGGUUUUAAGCCGCUUUUUAGCCCAAGUCGUUUU----UUUUUCA .....................................(((((((..((((((((((.....)))))))))).....)))))))...----....... ( -17.70) >consensus CAACCCAACCACUCCCCUACACCCCUUUUCCACGCUUGCGAUUUACGAGAAGCGGUUUUAAGCCGCUUUUUAGC_CAAGUCGUUUUU_UUUUUUUCA .....................................(((((((..((((((((((.....)))))))))).....))))))).............. (-16.33 = -16.67 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:00 2006