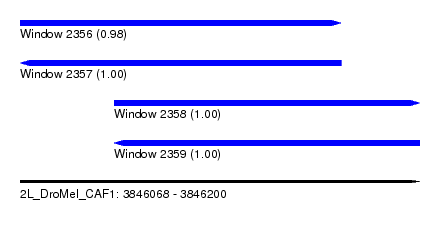
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,846,068 – 3,846,200 |
| Length | 132 |
| Max. P | 0.998649 |

| Location | 3,846,068 – 3,846,174 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979508 |
| Prediction | RNA |
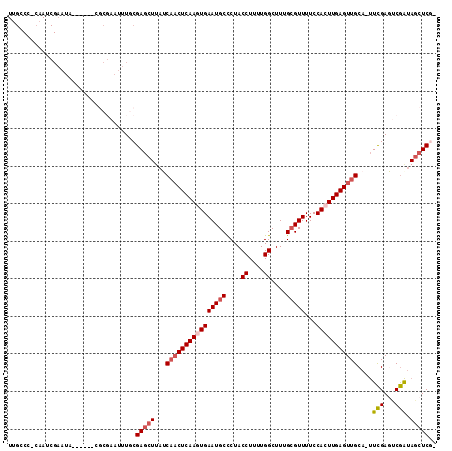
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3846068 106 + 22407834 UUGCCCGCAAACGAAUA------CGCGAAUUUGCGAGCUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCA-UUCGAGUCGAUAGCUCC- ..((.((((((((....------..))..)))))).))....((((((((((((((((....((....))....)))))...)))))))))))..-...((((.....)))).- ( -32.10) >DroSec_CAF1 34862 106 + 1 UAGCCCACAAUCGAAUA------CGCGAAUUUGCGAGCUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCA-UUCGAGUCGGUAGCUCG- ..(((.((..((((((.------((((....)))).......((((((((((((((((....((....))....)))))...))))))))))).)-))))))).)))......- ( -33.10) >DroSim_CAF1 34165 106 + 1 UUGCCAACAAUCGAAUA------CGCGAAUUUGCGAGCUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCA-UUCGAGUCGAUAGCUCG- ..((......((((((.------((((....)))).......((((((((((((((((....((....))....)))))...))))))))))).)-))))).......))...- ( -30.72) >DroWil_CAF1 65436 94 + 1 UUGCCG-GAAUCGAA-------A------GAUUCGA----AACUUCUCAAUUGAAUGCUCUACCUUUUGGCUUUGGGUUUUCCACUUGAGUUGCAAUUUGAUUCAA--ACUCCU ((((..-(((((...-------.------)))))..----.....(((((..(((.((((..((....))....)))).)))...)))))..))))..........--...... ( -21.10) >DroYak_CAF1 36768 93 + 1 -------------AAUAUCCAAA------UUUGCGAGCUUAACAACUCAACUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCA-UUCGAUGCGGUAGCUCG- -------------..........------....((((((...((((((((..((((((....((....))....)))))).....)))))))(((-(...)))))..))))))- ( -23.40) >consensus UUGCCC_CAAUCGAAUA______CGCGAAUUUGCGAGCUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCA_UUCGAGUCGAUAGCUCG_ ..................................(((((...((((((((((((((((....((....))....)))))...)))))))))))....(((...))).))))).. (-19.91 = -20.80 + 0.89)

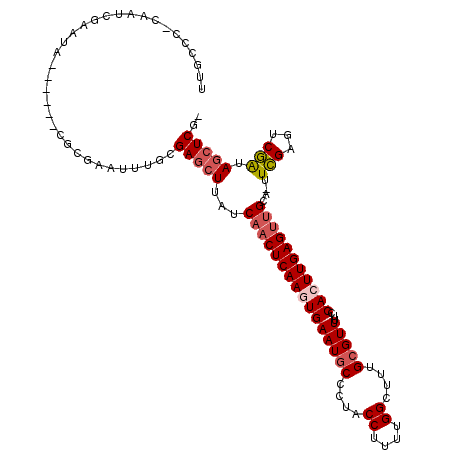
| Location | 3,846,068 – 3,846,174 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -21.37 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3846068 106 - 22407834 -GGAGCUAUCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAGCUCGCAAAUUCGCG------UAUUCGUUUGCGGGCAA -.((((....).)))...-..(((((((((((((....((...(((....)))...)).)))))))))))))....((((((((....(((------....))))))))))).. ( -38.20) >DroSec_CAF1 34862 106 - 1 -CGAGCUACCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAGCUCGCAAAUUCGCG------UAUUCGAUUGUGGGCUA -(((((....).))))..-..(((((((((((((....((...(((....)))...)).)))))))))))))...(((((((((..(((..------....)))))))))))). ( -38.70) >DroSim_CAF1 34165 106 - 1 -CGAGCUAUCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAGCUCGCAAAUUCGCG------UAUUCGAUUGUUGGCAA -...((((.(((.(((((-(((((((((((((((....((...(((....)))...)).)))))))))))).........((......)))------)))))))))).)))).. ( -38.80) >DroWil_CAF1 65436 94 - 1 AGGAGU--UUGAAUCAAAUUGCAACUCAAGUGGAAAACCCAAAGCCAAAAGGUAGAGCAUUCAAUUGAGAAGUU----UCGAAUC------U-------UUCGAUUC-CGGCAA .(((((--(.((.(((((((....(((((.(((.....)))..(((....)))...........))))).))))----).)).))------.-------...)))))-)..... ( -17.40) >DroYak_CAF1 36768 93 - 1 -CGAGCUACCGCAUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCAGUUGAGUUGUUAAGCUCGCAAA------UUUGGAUAUU------------- -((((((...((((...)-)))(((((((.((((....((...(((....)))...)).)))).)))))))....))))))....------..........------------- ( -27.00) >consensus _CGAGCUAUCGACUCGAA_UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAGCUCGCAAAUUCGCG______UAUUCGAUUG_GGGCAA ..(((((..((...)).....(((((((((((((....((...(((....)))...)).)))))))))))))...))))).................................. (-21.37 = -22.50 + 1.13)

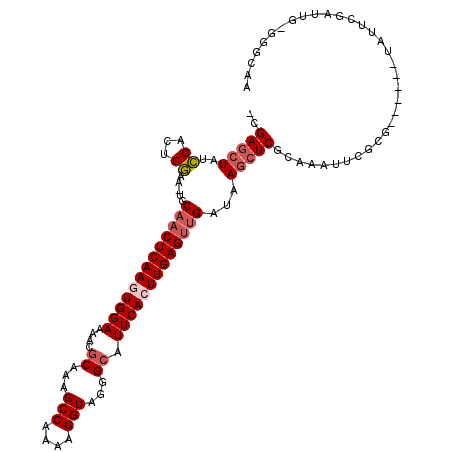
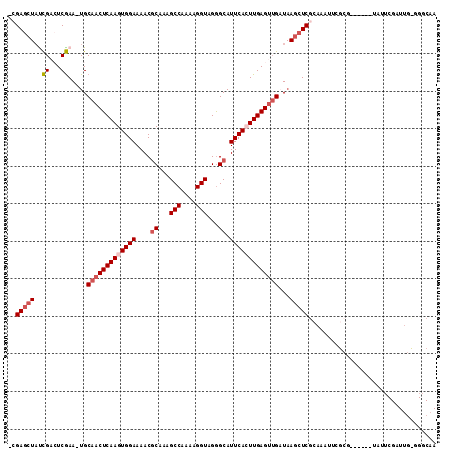
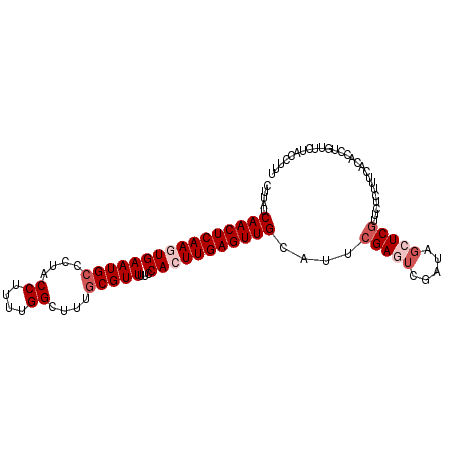
| Location | 3,846,099 – 3,846,200 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.76 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3846099 101 + 22407834 CUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCAUUCGAGUCGAUAGCUCCCUCUCUUUCACACCUGUUCUACCUUU .....((((((((((((((((....((....))....)))))...))))))))))).....((((.....))))........................... ( -23.10) >DroSec_CAF1 34893 101 + 1 CUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCAUUCGAGUCGGUAGCUCGUUCUCUUUCACACCUGUUCUACCUCU .....((((((((((((((((....((....))....)))))...)))))))))))..........(((((..((..............))..)))))... ( -25.34) >DroSim_CAF1 34196 101 + 1 CUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCAUUCGAGUCGAUAGCUCGUUCUCUUUCACACCUGUUCUACCUCU .....((((((((((((((((....((....))....)))))...)))))))))))....(((((.....))))).......................... ( -25.10) >DroEre_CAF1 39841 88 + 1 CUCAUCAACUCAACUGAAUGCUCUA-------------CGUUUUCCACUUGAGUUGCAUUCGAUUCGCUAGCUCGUUCUCUUUCACACCUGUUCUACCUUU .....((((((((..(((.((....-------------.)).)))...))))))))............................................. ( -10.10) >DroYak_CAF1 36786 101 + 1 CUUAACAACUCAACUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCAUUCGAUGCGGUAGCUCGUACUUUUUCAUACCUGUUCUACCUUU .....((((((((..((((((....((....))....)))))).....))))))))..........(((((..((((........)))..)..)))))... ( -20.30) >consensus CUUAUCAACUCAAGUGAAUGCCCUACCUUUUGGCUUUGCGUUUUCCACUUGAGUUGCAUUCGAGUCGAUAGCUCGUUCUCUUUCACACCUGUUCUACCUUU .....((((((((((((((((....((....))....)))))...)))))))))))....(((((.....))))).......................... (-17.26 = -18.76 + 1.50)

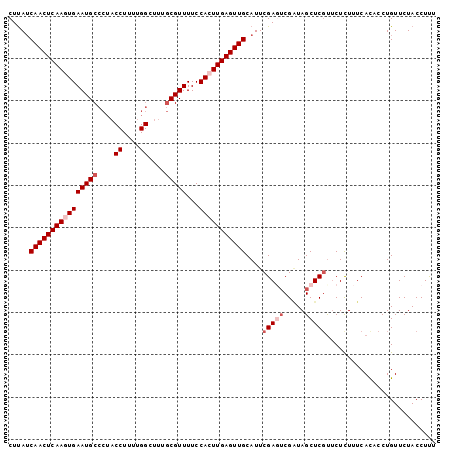
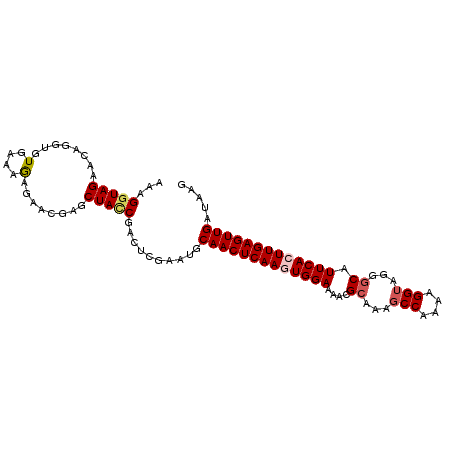
| Location | 3,846,099 – 3,846,200 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3846099 101 - 22407834 AAAGGUAGAACAGGUGUGAAAGAGAGGGAGCUAUCGACUCGAAUGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAG ...(((((..(.....(....).....)..)))))..........(((((((((((((....((...(((....)))...)).)))))))))))))..... ( -27.00) >DroSec_CAF1 34893 101 - 1 AGAGGUAGAACAGGUGUGAAAGAGAACGAGCUACCGACUCGAAUGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAG ...(((((......(((........)))..)))))..........(((((((((((((....((...(((....)))...)).)))))))))))))..... ( -30.10) >DroSim_CAF1 34196 101 - 1 AGAGGUAGAACAGGUGUGAAAGAGAACGAGCUAUCGACUCGAAUGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAG .(((...((.....(((........))).....))..))).....(((((((((((((....((...(((....)))...)).)))))))))))))..... ( -28.00) >DroEre_CAF1 39841 88 - 1 AAAGGUAGAACAGGUGUGAAAGAGAACGAGCUAGCGAAUCGAAUGCAACUCAAGUGGAAAACG-------------UAGAGCAUUCAGUUGAGUUGAUGAG ...(.(((......(((........)))..))).)...(((....((((((((.((((.....-------------.......)))).)))))))).))). ( -16.00) >DroYak_CAF1 36786 101 - 1 AAAGGUAGAACAGGUAUGAAAAAGUACGAGCUACCGCAUCGAAUGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCAGUUGAGUUGUUAAG ...(((((.....((((......))))...))))).........(((((((((.((((....((...(((....)))...)).)))).))))))))).... ( -28.70) >consensus AAAGGUAGAACAGGUGUGAAAGAGAACGAGCUACCGACUCGAAUGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAUAAG ...(((((........(....)........)))))..........(((((((((((((....((...(((....)))...)).)))))))))))))..... (-21.67 = -22.45 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:56 2006