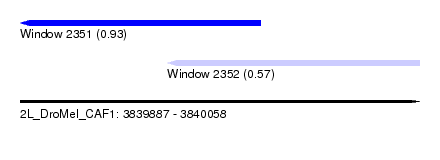
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,839,887 – 3,840,058 |
| Length | 171 |
| Max. P | 0.928167 |

| Location | 3,839,887 – 3,839,990 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
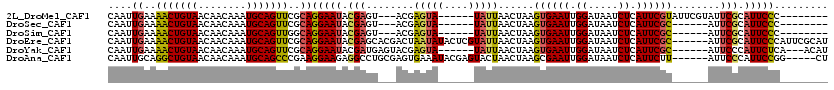
>2L_DroMel_CAF1 3839887 103 - 22407834 CAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUACGAGU---ACGAGUA------UAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGUAUUCGUAUUCGCAUUCCC-------- ...(((..(((((((........)))))))..)))((((((((((---((((((.------.((((.((((.....)))).))))...))))))))))))))))........-------- ( -33.20) >DroSec_CAF1 28704 97 - 1 CAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUACGAGU---ACGAGUA------UAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGC------AUUCGCAUUCCC-------- ....((..(((((((........)))))))..))(((((.(((((---.(((((.------.((((.((((.....)))).))))...))))).------))))).))))).-------- ( -25.90) >DroSim_CAF1 28027 97 - 1 CAAUUGAAAACUGUAACAACAAAUGCAGUUGGCAGGAAUACGAGU---ACGAGUA------UAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGC------AUUCGCAUUCCC-------- ....((..(((((((........)))))))..))(((((.(((((---.(((((.------.((((.((((.....)))).))))...))))).------))))).))))).-------- ( -26.50) >DroEre_CAF1 33384 114 - 1 CAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUACGAGCACGACUAAUAUACUCGUAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGC------AUUCGCAUUCCCAUUCGCAU ....((..(((((((........)))))))..))((((((((((.............))))))))......(((((((((((......))))).------))))))....))........ ( -25.02) >DroYak_CAF1 30157 105 - 1 CAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUACGAUGAGUACGAGUA------UAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGC------AUUCCCAUUCUCA---ACAU ....((..(((((((........)))))))..))(((((.(((((((........------.((((.((((.....)))).)))))))).))).------)))))........---.... ( -20.50) >DroAna_CAF1 29887 109 - 1 CAAUUGCAGGCUGUAACAACAAAUGCAGCCCGAAGGAAGAGGCCUGCGAGUGAAAUACGAGUACUAACUAAGCGAAUUGGAUAAUCUCAUUCUU------AUUCCCAUUCCGG-----CU .....((.(((((((........)))))))....((((..((..((.((((((..((....))....((((.....))))......)))))).)------)..))..)))).)-----). ( -24.50) >consensus CAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUACGAGU___ACGAGUA______UAUUAACUAAGUGAAUUGGAUAAUCUCAUUCGC______AUUCGCAUUCCC________ ....((..(((((((........)))))))..))(((((.(((........(((((....)))))......((((((.((.....)).))))))........))).)))))......... (-17.00 = -16.95 + -0.05)

| Location | 3,839,950 – 3,840,058 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.49 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3839950 108 - 22407834 AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUA .......((..((((.......(((.((....))((.(((..(......)..))).)).)))......))))..))(((((((........))))))).......... ( -19.12) >DroSec_CAF1 28761 108 - 1 AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUA .......((..((((.......(((.((....))((.(((..(......)..))).)).)))......))))..))(((((((........))))))).......... ( -19.12) >DroSim_CAF1 28084 108 - 1 AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUGGCAGGAAUA .......((..((((.......(((.((....))((.(((..(......)..))).)).)))......))))..))(((((((........))))))).......... ( -19.72) >DroEre_CAF1 33458 108 - 1 AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUA .......((..((((.......(((.((....))((.(((..(......)..))).)).)))......))))..))(((((((........))))))).......... ( -19.12) >DroYak_CAF1 30222 108 - 1 AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUA .......((..((((.......(((.((....))((.(((..(......)..))).)).)))......))))..))(((((((........))))))).......... ( -19.12) >DroAna_CAF1 29956 108 - 1 AAUUUACUUUGAUUGAAUAAUGGCCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGCAGGCUGUAACAACAAAUGCAGCCCGAAGGAAGA ......(((((((((.......(((.((....))((.(((..(......)..))).)).)))......))))....(((((((........))))))))))))..... ( -26.62) >consensus AAUUUACUUUGAUUGAAUAAUGGUCGCAUAAUUGGCAACUUGAAUGGAAUUGAGUAGCAGGCAAAACGCAAUUGAAAACUGUAACAACAAAUGCAGUUCGCAGGAAUA .......((((((((.......(((.((....))((.(((..(......)..))).)).)))......))))))))(((((((........))))))).......... (-20.01 = -19.49 + -0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:50 2006