
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,838,278 – 3,838,386 |
| Length | 108 |
| Max. P | 0.825234 |

| Location | 3,838,278 – 3,838,386 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -24.94 |
| Energy contribution | -26.17 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3838278 108 + 22407834 CACCGCC----CUU--------CGUCGCACCACCCACGAGCAGUGAGCUCUACCAGCGAUUGCAGUACCAACAGCUGUUGCAUCAACAUGCGGCAGCCGCCGCUCUGGCGGCUCAUCAAA .......----...--------.((((((........((((.....)))).....(((((.((.((....)).)).))))).......))))))(((((((.....)))))))....... ( -35.90) >DroSec_CAF1 27146 108 + 1 CUCCACC----CUU--------UGUUGCACCACCCACGAGCAGUGAACUCUACCAGCGAUUGCAGUACCAACAGCUGUUGCAUCAACAUGCGGCAGCCGCCGCCCUGGCGGCUCAUCAAA .......----...--------(((((.......(((.....)))..........((....)).....)))))((((((((((....)))))))))).(((((....)))))........ ( -32.40) >DroEre_CAF1 31737 108 + 1 CGCCGCC----CUU--------CGUUGCACCACCCACGAGCAGUGAGCUCUACCAGCGAUUGCAGUACCAACAGCUGUUGCACCAACAUGCGGCAGCCGCCGCACUGGCGGCUCAUCAAA .((((((----...--------.((((..........((((.....)))).....(((((.((.((....)).)).)))))..)))).((((((....))))))..))))))........ ( -40.60) >DroWil_CAF1 51238 120 + 1 CGCCAUCAGUCCAACCUAAUCCCGCCGCCGCCGCCGCAAGCACGGAACUGUAUCAGCGUCUGCAGUAUCAACAGUUGUUGCAUCAACAUGCAGCGGCAGCCGCUUUGGCGGCACAUCAAA .......................(((((((..((.((......((.((((((........)))))).))....((((((((((....)))))))))).)).))..)))))))........ ( -40.10) >DroYak_CAF1 28509 108 + 1 CUCCGCC----CUU--------UGUUGCACCGCCCACGAGCAGUGAGCUCUACCAGCGAUUGCAGUACCAACAGCUGUUGCAUCAACAUGCGGCAGCCGCCGCAUUGGCGGCACAUCAAA ..(((((----..(--------((.((((.(((....((((.....)))).....)))...(((((.......))))))))).))).(((((((....))))))).)))))......... ( -37.70) >DroAna_CAF1 28393 104 + 1 -UCCGCC----CUU--------CGUGACCCCGCCCACGAGC---GAGCUCUACCAGCGCCUGCAGUACCAGCAGCUCCUGCACCAGCACGCGGCCGCCGCGGCUCUGGCGGCCCACCAGC -......----..(--------((((........)))))((---(.(((....(((...((((.......))))...)))....))).)))((((((((......))))))))....... ( -37.10) >consensus CUCCGCC____CUU________CGUUGCACCACCCACGAGCAGUGAGCUCUACCAGCGAUUGCAGUACCAACAGCUGUUGCAUCAACAUGCGGCAGCCGCCGCUCUGGCGGCUCAUCAAA .......................(((((.........((((.....)))).....(((((.((.((....)).)).)))))........))))).((((((.....))))))........ (-24.94 = -26.17 + 1.22)


| Location | 3,838,278 – 3,838,386 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -47.53 |
| Consensus MFE | -33.49 |
| Energy contribution | -34.80 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
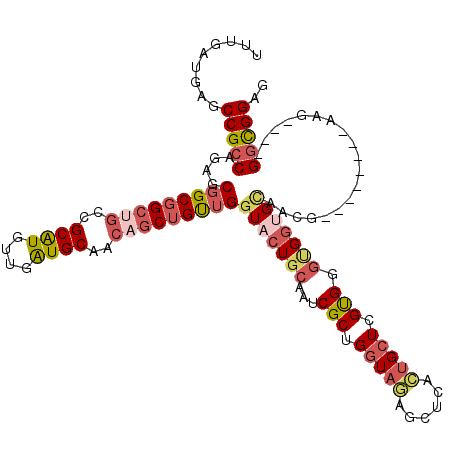
>2L_DroMel_CAF1 3838278 108 - 22407834 UUUGAUGAGCCGCCAGAGCGGCGGCUGCCGCAUGUUGAUGCAACAGCUGUUGGUACUGCAAUCGCUGGUAGAGCUCACUGCUCGUGGGUGGUGCGACG--------AAG----GGCGGUG ........((((((....(((((((((..((((....))))..)))))))))((((..(...(((.(((((......))))).))).)..))))....--------...----)))))). ( -46.10) >DroSec_CAF1 27146 108 - 1 UUUGAUGAGCCGCCAGGGCGGCGGCUGCCGCAUGUUGAUGCAACAGCUGUUGGUACUGCAAUCGCUGGUAGAGUUCACUGCUCGUGGGUGGUGCAACA--------AAG----GGUGGAG .........(((((....(((((((((..((((....))))..)))))))))((((..(...(((.(((((......))))).))).)..))))....--------...----))))).. ( -42.50) >DroEre_CAF1 31737 108 - 1 UUUGAUGAGCCGCCAGUGCGGCGGCUGCCGCAUGUUGGUGCAACAGCUGUUGGUACUGCAAUCGCUGGUAGAGCUCACUGCUCGUGGGUGGUGCAACG--------AAG----GGCGGCG ........(((((((((((((((((((..((((....))))..)))))))).)))))(((.(((((....((((.....))))...))))))))....--------...----)))))). ( -49.20) >DroWil_CAF1 51238 120 - 1 UUUGAUGUGCCGCCAAAGCGGCUGCCGCUGCAUGUUGAUGCAACAACUGUUGAUACUGCAGACGCUGAUACAGUUCCGUGCUUGCGGCGGCGGCGGCGGGAUUAGGUUGGACUGAUGGCG ........(((.((...((.((((((((((((.......(((..((((((.......((....))....))))))...))).)))))))))))).)).))(((((......)))))))). ( -49.60) >DroYak_CAF1 28509 108 - 1 UUUGAUGUGCCGCCAAUGCGGCGGCUGCCGCAUGUUGAUGCAACAGCUGUUGGUACUGCAAUCGCUGGUAGAGCUCACUGCUCGUGGGCGGUGCAACA--------AAG----GGCGGAG .........(((((....(((((((((..((((....))))..)))))))))(((((((...(((.(((((......))))).))).)))))))....--------...----))))).. ( -46.90) >DroAna_CAF1 28393 104 - 1 GCUGGUGGGCCGCCAGAGCCGCGGCGGCCGCGUGCUGGUGCAGGAGCUGCUGGUACUGCAGGCGCUGGUAGAGCUC---GCUCGUGGGCGGGGUCACG--------AAG----GGCGGA- ((..((((((((((........)))))))((((((..((((....)).))..)))).))...)))..)).((.(((---(((....)))))).)).(.--------...----).....- ( -50.90) >consensus UUUGAUGAGCCGCCAGAGCGGCGGCUGCCGCAUGUUGAUGCAACAGCUGUUGGUACUGCAAUCGCUGGUAGAGCUCACUGCUCGUGGGUGGUGCAACG________AAG____GGCGGAG .........(((((....(((((((((..((((....))))..)))))))))(((((((...(((.(((((......))))).))).)))))))...................))))).. (-33.49 = -34.80 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:47 2006