| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,833,164 – 3,833,267 |
| Length | 103 |
| Max. P | 0.817726 |

| Location | 3,833,164 – 3,833,267 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3833164 103 + 22407834 GGCUCAUCACCCCGAUGUUUGGGCAAACCAUAUGCUCU--GGCAGCAUGAACCAGUGUGUCCACGGUCCAUGGAGCAACCCCCAAAAAACGGACACCCCUUGCCC (((........(((...((((((.........(((...--.))).((((.(((.(((....)))))).))))........))))))...))).........))). ( -27.13) >DroSec_CAF1 22072 102 + 1 GGCUCAUCACCCCGAUGUUUGGGCAAACCAUAUGCUCU--GGCAGCAUGGACCAGUGUGUCCACGGUCCAUGGAGCAACCCCCAAAAAA-GGACACCCCUUGCCC (((.((((.....))))((((((.........(((...--.))).((((((((.(((....)))))))))))........)))))).((-((.....))))))). ( -34.30) >DroEre_CAF1 26532 104 + 1 GGCUCAUCACCCCGAUGUUUGGGCAAACCAUAUGCUCU-GGGCAGCACGGACCAGUGUGUCCACGGUCCAUGGAGCAACCCCCAAAAAAAGGACACCCCUGGCCC ((((((((.....))))((((((....((((..(((.(-(((((.(((......))))))))).)))..)))).......))))))...(((.....))))))). ( -33.20) >DroYak_CAF1 23349 105 + 1 GGCUCAUCACCCCGAUGUUUGGGCAAACCAUAUGCUCUCUGGCAACAUGGACCAGUGUGUCCACGGUCCAUGGAGCAACCCCCAAAAAAAGGACACCCCCUGCCA (((.((((.....))))((((((.........((((....)))).((((((((.(((....)))))))))))........))))))...(((......)))))). ( -35.10) >consensus GGCUCAUCACCCCGAUGUUUGGGCAAACCAUAUGCUCU__GGCAGCAUGGACCAGUGUGUCCACGGUCCAUGGAGCAACCCCCAAAAAAAGGACACCCCUUGCCC (((.((((.....))))((((((.........((((....)))).((((((((.(((....)))))))))))........))))))....((.....))..))). (-30.38 = -30.87 + 0.50)



| Location | 3,833,164 – 3,833,267 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -36.62 |
| Energy contribution | -37.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3833164 103 - 22407834 GGGCAAGGGGUGUCCGUUUUUUGGGGGUUGCUCCAUGGACCGUGGACACACUGGUUCAUGCUGCC--AGAGCAUAUGGUUUGCCCAAACAUCGGGGUGAUGAGCC .(((...(((((.((((.((((((.((......(((((((((((....))).)))))))))).))--))))...))))..)))))...((((.....)))).))) ( -37.80) >DroSec_CAF1 22072 102 - 1 GGGCAAGGGGUGUCC-UUUUUUGGGGGUUGCUCCAUGGACCGUGGACACACUGGUCCAUGCUGCC--AGAGCAUAUGGUUUGCCCAAACAUCGGGGUGAUGAGCC ((((((((((((.((-((.....)))).))))))((((((((((....))).)))))))...(((--(.......))))))))))...((((.....)))).... ( -41.20) >DroEre_CAF1 26532 104 - 1 GGGCCAGGGGUGUCCUUUUUUUGGGGGUUGCUCCAUGGACCGUGGACACACUGGUCCGUGCUGCCC-AGAGCAUAUGGUUUGCCCAAACAUCGGGGUGAUGAGCC (((((((..((((((..((..(((((....)))))..))....)))))).)))))))(((((....-..)))))..(((((((((........))))...))))) ( -43.90) >DroYak_CAF1 23349 105 - 1 UGGCAGGGGGUGUCCUUUUUUUGGGGGUUGCUCCAUGGACCGUGGACACACUGGUCCAUGUUGCCAGAGAGCAUAUGGUUUGCCCAAACAUCGGGGUGAUGAGCC ((((((((((((.(((((....))))).))))))((((((((((....))).))))))).))))))..........(((((((((........))))...))))) ( -42.00) >consensus GGGCAAGGGGUGUCCUUUUUUUGGGGGUUGCUCCAUGGACCGUGGACACACUGGUCCAUGCUGCC__AGAGCAUAUGGUUUGCCCAAACAUCGGGGUGAUGAGCC (((((.((((((.(((((....))))).))))))((((((((((....))).)))))))..)))))..........(((((((((........))))...))))) (-36.62 = -37.38 + 0.75)

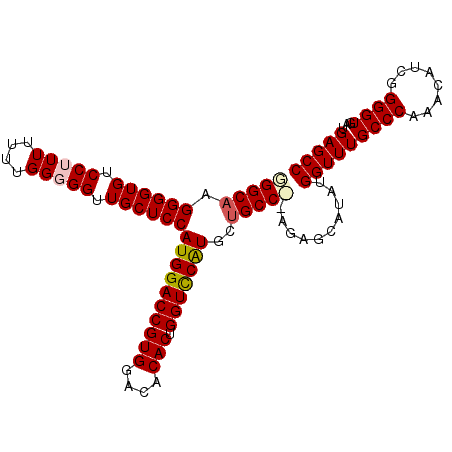

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:42 2006