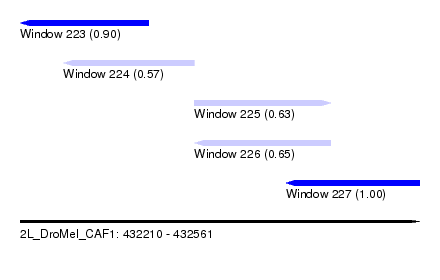
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 432,210 – 432,561 |
| Length | 351 |
| Max. P | 0.999484 |

| Location | 432,210 – 432,323 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 432210 113 - 22407834 AAAAAAAAAAAUUUAAAGAAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUAUGGUUGACAGAUAAGAAAUUUGCUGGCAUUUGUUUUGU ...............((((.(((.((..((((...((((((..((.....)).))))))...((..((.((...)).))..))...........))))..)).))).)))).. ( -23.40) >DroSim_CAF1 8750 108 - 1 AAAA---AAAAUUUAAA--AAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUAUGGUUGACAGAUAAGAAAUUUGCUGGCAUUUGUUUUGU ....---......((((--(.(((((..((((...((((((..((.....)).))))))...((..((.((...)).))..))...........))))..)).))).))))). ( -23.50) >DroEre_CAF1 9575 108 - 1 AAAA---AAGG--AAUGAAAAAAACCUGGCGACAACAACGAAAUGUUAAACAAUCGUCGGACGUUGGCGAUUUUAUGGUUGGCAGAUAAGAAAUUUGCUGGCAUUUGUUUUGU ....---.(((--...........)))....((((.((((((.........((((((((.....)))))))).....((..((((((.....))))))..)).)))))))))) ( -22.90) >DroYak_CAF1 9177 110 - 1 AAAA---AAGUUUAAAACAAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUAUGGUUGGCAGAUAAGAAAUUUGCUGGCAUUUGUUUUGU ....---.....(((((((((...((..(((....((((((..((.....)).))))))..)))..)).........((..((((((.....))))))..)).))))))))). ( -33.50) >consensus AAAA___AAAAUUAAAA_AAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUAUGGUUGACAGAUAAGAAAUUUGCUGGCAUUUGUUUUGU ........................((..((((...((((((..((.....)).))))))...((..((.((...)).))..))...........))))..))........... (-20.70 = -20.95 + 0.25)

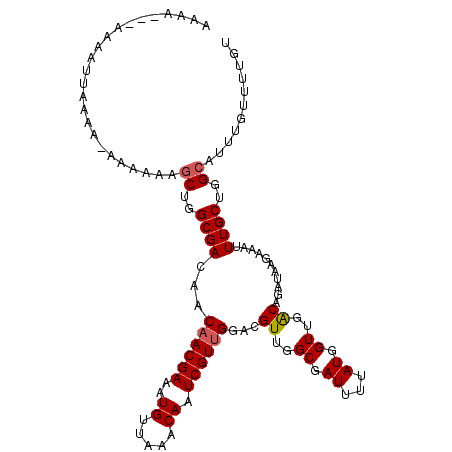
| Location | 432,248 – 432,363 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -22.15 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 432248 115 - 22407834 AUUGCACAAUUCGCUGAAUUUUUGACUCUUAGCGGGACGCAAAAAAAAAAAUUUAAAGAAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUA .((((....((((((((...........))))))))..))))................((((..((..(((....((((((..((.....)).))))))..)))..))..)))). ( -26.10) >DroSim_CAF1 8788 110 - 1 AUUGCACAAUUCGCUGGAUUUUCGACUCUUAGCGGGACGCAAAA---AAAAUUUAAA--AAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUA .((((....(((((((((........)).)))))))..))))..---.........(--(((..((..(((....((((((..((.....)).))))))..)))..))..)))). ( -26.20) >DroEre_CAF1 9613 110 - 1 AUUGCACAAUUCGCUGGAUUUGCGACUGUUAGCGGGGCGCAAAA---AAGG--AAUGAAAAAAACCUGGCGACAACAACGAAAUGUUAAACAAUCGUCGGACGUUGGCGAUUUUA (((((.....((((..(.((((((.(((....)))..)))))).---....--..(....)....)..))))((((..(((..((.....))....)))...))))))))).... ( -26.90) >DroYak_CAF1 9215 111 - 1 AUUGCACAAUUCGCUGGAUUUUCGACUGUUGGCG-GGCGCAAAA---AAGUUUAAAACAAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUA (((((.((((((((..(.((((.((((....((.-...))....---.)))).))))........)..))))...((((((..((.....)).))))))...))))))))).... ( -24.90) >consensus AUUGCACAAUUCGCUGGAUUUUCGACUCUUAGCGGGACGCAAAA___AAAAUUAAAA_AAAAAAGCUGGCGACAACAACGAAAUGUUAAACAAUCGUUGGACGUUGGCGAUUUUA .((((....((((((((...........))))))))..))))......................((..(((....((((((..((.....)).))))))..)))..))....... (-22.15 = -22.52 + 0.38)

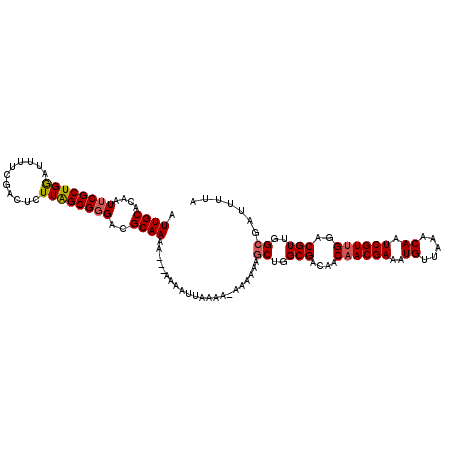
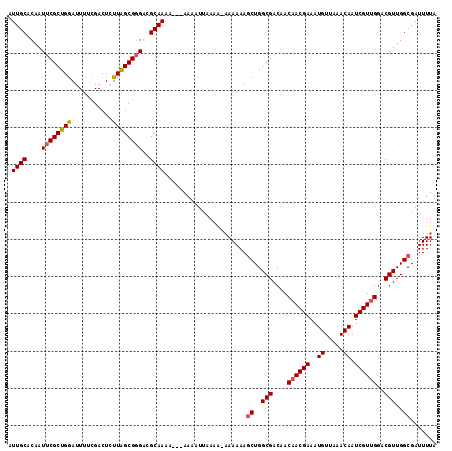
| Location | 432,363 – 432,483 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.99 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -19.58 |
| Energy contribution | -22.48 |
| Covariance contribution | 2.90 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 432363 120 + 22407834 UAACCCAUUUCGGGCCAUGGAAUAUCAAAUGUCAGUCAAACUUGACACUCUACGGAGGGUAUAUUGAAAUGGGGAAGCUGUUUAGAGUACUAUUGUGUCAAAUAUAUAUGUACAUUCACA ...(((((((((.(((.............((((((......))))))(((....))))))....))))))))).............((((...(((((...)))))...))))....... ( -31.00) >DroSec_CAF1 9910 120 + 1 UAACCCAUUUCGGGCCAUGGAAUAUCAAAUGUCAGUCAAACUUGACACUCUACGGAGCGUAUAUUGAAAUGGGGAAGCUGUUUAGAGUAAUAUUUUGUCAUUUAAAUAUGCACAUUCAUA ...((((((((((...(((..........((((((......))))))(((....))))))...))))))))))...((((((((((((((....))).).)))))))).))......... ( -29.20) >DroSim_CAF1 8898 120 + 1 UAACCCAUUUCGGGCCAUGGAAUAUCAAAUGUCAGUCAAACUUGACACUCCACGGAGGGUAUAUUGAAAUGGGGAAGCUGUUUAGAGUAAUAUUUUGUCAUUUAAAUAUGCACAUUAAUA ...(((((((((.(((.............((((((......))))))(((....))))))....)))))))))...((((((((((((((....))).).)))))))).))......... ( -30.10) >DroEre_CAF1 9723 119 + 1 UAACCCAUUUCUGGCCAUGGAAUAUCAAAUGUCAGUCAAAUUUGACACUCUUCGGAGGGUAUAUUGAAAUAGGGAAGCUGUUUUGGG-AAUAUUUUGUCAUUUAUAUACAUAUAUACCUA ...((((((((..(((.............((((((......))))))(((....)))))).....))))).))).........((((-.((((..(((.((....))))).)))).)))) ( -26.90) >DroYak_CAF1 9326 101 + 1 UAACCCAUUUCGGGCCAUGGAAUAUCAAAUGUCAGUCAAAUUUGACACUCUACGGAGGGUAUAUUGAAAUAGGGAAGCUGUUU-----------UUCUCAUUUAUAUAUAUA-------- ...(((((((((.(((.............((((((......))))))(((....))))))....)))))).))).........-----------..................-------- ( -22.90) >consensus UAACCCAUUUCGGGCCAUGGAAUAUCAAAUGUCAGUCAAACUUGACACUCUACGGAGGGUAUAUUGAAAUGGGGAAGCUGUUUAGAGUAAUAUUUUGUCAUUUAUAUAUGUACAUUCAUA ...(((((((((.(((.............((((((......))))))(((....))))))....)))))))))............................................... (-19.58 = -22.48 + 2.90)

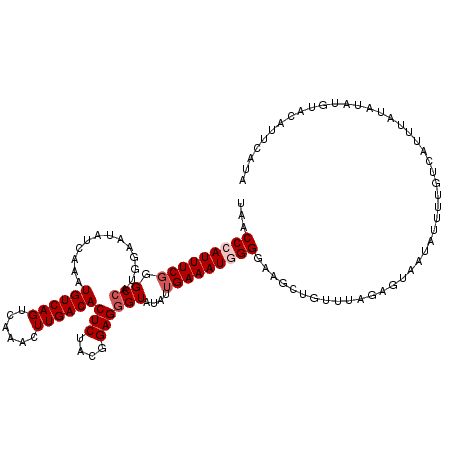
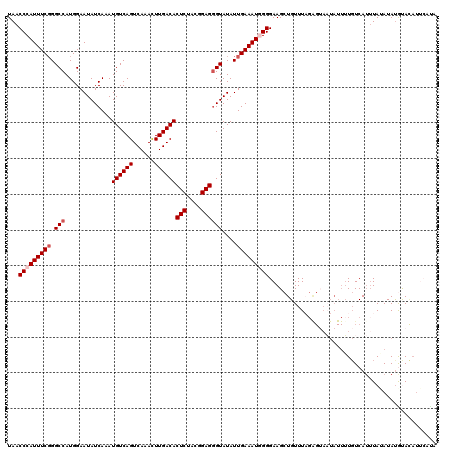
| Location | 432,363 – 432,483 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.99 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 432363 120 - 22407834 UGUGAAUGUACAUAUAUAUUUGACACAAUAGUACUCUAAACAGCUUCCCCAUUUCAAUAUACCCUCCGUAGAGUGUCAAGUUUGACUGACAUUUGAUAUUCCAUGGCCCGAAAUGGGUUA ((((((((((....))))))...))))....................((((((((..........((((.(((((((((((.........))))))))))).))))...))))))))... ( -25.22) >DroSec_CAF1 9910 120 - 1 UAUGAAUGUGCAUAUUUAAAUGACAAAAUAUUACUCUAAACAGCUUCCCCAUUUCAAUAUACGCUCCGUAGAGUGUCAAGUUUGACUGACAUUUGAUAUUCCAUGGCCCGAAAUGGGUUA .......(((.((((((........))))))))).............((((((((.......(..((((.(((((((((((.........))))))))))).))))..)))))))))... ( -24.31) >DroSim_CAF1 8898 120 - 1 UAUUAAUGUGCAUAUUUAAAUGACAAAAUAUUACUCUAAACAGCUUCCCCAUUUCAAUAUACCCUCCGUGGAGUGUCAAGUUUGACUGACAUUUGAUAUUCCAUGGCCCGAAAUGGGUUA ..(((..(((.((((((........)))))))))..)))........((((((((..........((((((((((((((((.........))))))))))))))))...))))))))... ( -31.22) >DroEre_CAF1 9723 119 - 1 UAGGUAUAUAUGUAUAUAAAUGACAAAAUAUU-CCCAAAACAGCUUCCCUAUUUCAAUAUACCCUCCGAAGAGUGUCAAAUUUGACUGACAUUUGAUAUUCCAUGGCCAGAAAUGGGUUA ..(((((((.((...(((..............-................)))..)))))))))..(((..(((((((((((.........)))))))))))..)))(((....))).... ( -19.71) >DroYak_CAF1 9326 101 - 1 --------UAUAUAUAUAAAUGAGAA-----------AAACAGCUUCCCUAUUUCAAUAUACCCUCCGUAGAGUGUCAAAUUUGACUGACAUUUGAUAUUCCAUGGCCCGAAAUGGGUUA --------..................-----------..........((((((((..........((((.(((((((((((.........))))))))))).))))...))))))))... ( -19.72) >consensus UAUGAAUGUACAUAUAUAAAUGACAAAAUAUUACUCUAAACAGCUUCCCCAUUUCAAUAUACCCUCCGUAGAGUGUCAAGUUUGACUGACAUUUGAUAUUCCAUGGCCCGAAAUGGGUUA ...............................................((((((((..........((((.(((((((((((.........))))))))))).))))...))))))))... (-21.10 = -20.82 + -0.28)

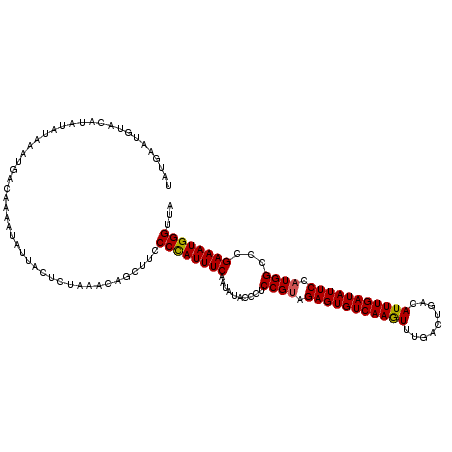
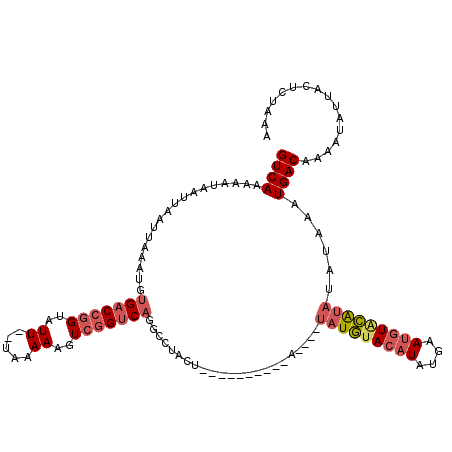
| Location | 432,443 – 432,561 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.52 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 432443 118 - 22407834 GUCAAAAAAAAUUAAUUAAAUGUGACCGGUAUU--UAAAAAGUCGGUCAGGCCUACUAUAUAACUACAUAUGUAUGCACAUGUGAAUGUACAUAUAUAUUUGACACAAUAGUACUCUAAA ((((((..............(.(((((((....--.......))))))).).................((((((((.((((....)))).))))))))))))))................ ( -24.40) >DroSec_CAF1 9990 95 - 1 GUCAAAACUAAUUAAUUAAAUGUGACCGGUAUU--UAAAAAGUCGGUC-------------------A----UAUGUACAUAUGAAUGUGCAUAUUUAAAUGACAAAAUAUUACUCUAAA ((((...............((((((((((....--.......))))))-------------------)----)))((((((....)))))).........))))................ ( -21.60) >DroSim_CAF1 8978 105 - 1 GUCAAAACUAAUUAAUUAAAUGUGACCGGUAUU--UAAAAAGUCGGUCAUGCCCACC---------CA----UAUGUACAUAUUAAUGUGCAUAUUUAAAUGACAAAAUAUUACUCUAAA ((((.................((((((((....--.......)))))))).......---------.(----(((((((((....)))))))))).....))))................ ( -23.20) >DroEre_CAF1 9803 103 - 1 GUCAAAAAUAACUAAUUAAAUAUGACCGGUAUUUUUAAAAACUACGACAGGUCUACUU----------------UAUACCUAGGUAUAUAUGUAUAUAAAUGACAAAAUAUU-CCCAAAA ((((...................((((.((................)).))))(((..----------------(((((....)))))...)))......))))........-....... ( -14.19) >consensus GUCAAAAAUAAUUAAUUAAAUGUGACCGGUAUU__UAAAAAGUCGGUCAGGCCUACU__________A____UAUGUACAUAUGAAUGUACAUAUAUAAAUGACAAAAUAUUACUCUAAA ((((..................(((((((..((.....))..))))))).......................(((((((((....)))))))))......))))................ (-12.24 = -13.80 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:22 2006