
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,825,330 – 3,825,434 |
| Length | 104 |
| Max. P | 0.947797 |

| Location | 3,825,330 – 3,825,434 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -15.81 |
| Energy contribution | -15.15 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
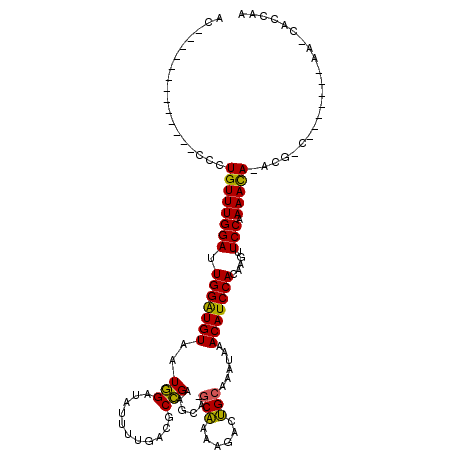
| Structure conservation index | 0.65 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
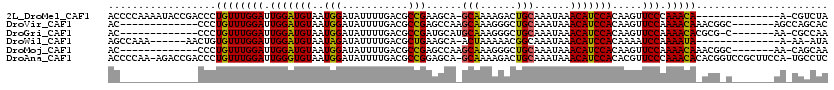
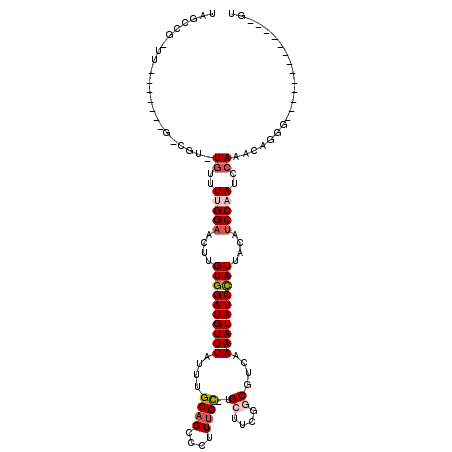
>2L_DroMel_CAF1 3825330 104 + 22407834 ACCCCAAAAUACCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA-GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA--------------A-CGUCUA .............(((..(((((((..(((((((..(((...........)))..(((-((....).))))......))))))).......)))))))--------------.-.))).. ( -24.00) >DroVir_CAF1 30820 100 + 1 AC-------------CCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAGCCAAGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACAAACGGC-------AGCCAGCAC ..-------------..(((((((..(((((.....(((((.(((....((..((((........))))))....))).))))).....)))))..))))))).-------......... ( -27.10) >DroGri_CAF1 18147 98 + 1 AC-------------CCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAUGCAUGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACACGCG-C-------AA-CGCCAA ..-------------...((((((((.(((((((..(((...........))).((((.((.....)))))).....))))))).....))).))))).(((-.-------..-)))... ( -21.50) >DroWil_CAF1 34974 97 + 1 AGCCAAA------AACUGUGUUUGGAUUGGAUGUAAUAGAUAUUUUGACGCUGAAGCA-ACUAAAAACGGCAAAUAAACAUCCACAAAAUCCAAAAUA--------------A-AA-AUA .......------.......((((((((((((((....((....))...((((.....-........))))......)))))))....)))))))...--------------.-..-... ( -16.22) >DroMoj_CAF1 20249 99 + 1 AC-------------CCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAGCCAAGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACAAACGGC-------AA-CAGCAA ..-------------..(((((((..(((((.....(((((.(((....((..((((........))))))....))).))))).....)))))..))))))).-------..-...... ( -27.10) >DroAna_CAF1 15870 117 + 1 ACCCCAA-AGACCGACCCUGUUUGGAUUGGGUGUAAUGGAUAUUUUGACGCCGGAGCA-GCAAAAGACUGCAAAUAAACAUCCACACGUUCCCAAACACACGGUCCGCUUCCA-UGCCUC .......-.(((((....(((((((...(..(((..(((((..((((....))))(((-((....).))))........))))).)))..))))))))..))))).((.....-.))... ( -29.30) >consensus AC_____________CCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA_GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCAAAACA_ACG_C_______AA_CACCAA ..................((((((((.(((((((..(((...........)))......(((......)))......))))))).....))).)))))...................... (-15.81 = -15.15 + -0.66)
| Location | 3,825,330 – 3,825,434 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
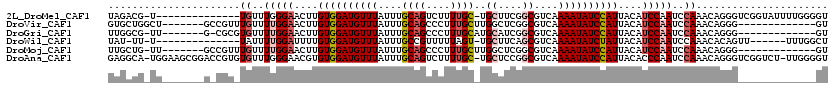
>2L_DroMel_CAF1 3825330 104 - 22407834 UAGACG-U--------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGGUAUUUUGGGGU ..(((.-(--------------((((((((.....((((((((((....((((....))))-.((....))....)))))))))).........))))))))).)))............. ( -33.04) >DroVir_CAF1 30820 100 - 1 GUGCUGGCU-------GCCGUUUGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCUUGGCUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGG-------------GU ......(((-------.(.(((((..(((((....((((((((((...(((((((........))))..)))...))))))))))....)))))..))))).).)-------------)) ( -34.40) >DroGri_CAF1 18147 98 - 1 UUGGCG-UU-------G-CGCGUGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCAUGCAUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGG-------------GU ((((((-((-------(-.((((((...((...(((..(((....)))..))).))...))))))..))))))))).................((((......))-------------)) ( -26.90) >DroWil_CAF1 34974 97 - 1 UAU-UU-U--------------UAUUUUGGAUUUUGUGGAUGUUUAUUUGCCGUUUUUAGU-UGCUUCAGCGUCAAAAUAUCUAUUACAUCCAAUCCAAACACAGUU------UUUGGCU ...-..-.--------------....((((((...((((((((((...(((.((.......-.))....)))...))))))))))...)))))).(((((.......------))))).. ( -18.10) >DroMoj_CAF1 20249 99 - 1 UUGCUG-UU-------GCCGUUUGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCUUGGCUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGG-------------GU ......-..-------.(((((((..(((((....((((((((((...(((((((........))))..)))...))))))))))....)))))..))))).)).-------------.. ( -32.70) >DroAna_CAF1 15870 117 - 1 GAGGCA-UGGAAGCGGACCGUGUGUUUGGGAACGUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUCCGGCGUCAAAAUAUCCAUUACACCCAAUCCAAACAGGGUCGGUCU-UUGGGGU ...((.-(.(((((.((((.(((..(((((.....((((((((((....((((....))))-.((....))....))))))))))....))))).....))).)))).).))-)).).)) ( -38.10) >consensus UAGCCG_UU_______G_CGU_UGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGC_UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGG_____________GU ......................((..(((((....((((((((((....((((....))))..((....))....))))))))))....)))))..))...................... (-16.34 = -17.23 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:39 2006