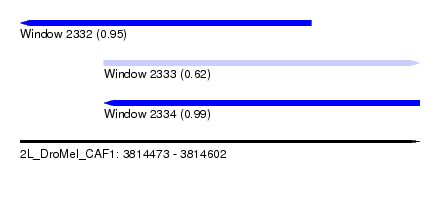
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,814,473 – 3,814,602 |
| Length | 129 |
| Max. P | 0.990755 |

| Location | 3,814,473 – 3,814,567 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -10.98 |
| Consensus MFE | -1.54 |
| Energy contribution | -3.18 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.14 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3814473 94 - 22407834 GCUCCACA--AAC-CAACCAAUUGGUUGGAACAUAAAGUUCAACAACAAACAGAGCCAUAAACAUUCCCAU-UACAAAUUAACACUUC------AUAUAUAUUC ((((....--...-(((((....)))))((((.....))))...........))))...............-................------.......... ( -14.50) >DroSim_CAF1 3478 94 - 1 GCUUCACA--AAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCAU-UACAAAUUAACACUUC------AUAUAUAUUC ((((....--(((-(((....))))))(((((.....)))))..........))))...............-................------.......... ( -13.70) >DroEre_CAF1 7747 96 - 1 GCGCCACAAGAAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCAU-UACAAAUUAACACUUC------AUAUAUAUUC ((........(((-(((....))))))(((((.....)))))............))...............-................------.......... ( -12.40) >DroWil_CAF1 13338 99 - 1 GCUUUUCA--UGCUGAACCAAUUUGUAGAGUCAUAAAAUUGAACAACAAACAU---CAUCAUCAUAAUCAUCAACAAAUUAACACUUCACAUUCAUAUAUGCUC .......(--((.(((.....(((((....(((......)))...)))))...---..))).)))....................................... ( -6.50) >DroAna_CAF1 4023 79 - 1 ----------GA--GCACCAAUUGGCUGGAACAUAAAGUUC---AACAAACAGAGCCAUAAACAUUCCC-C-UACAAAUUAACACUUC------AUA--UAUUC ----------..--........(((((.((((.....))))---..(.....))))))...........-.-................------...--..... ( -7.80) >consensus GCUCCACA__AAC_CAACCAAUUGGUUGGAACAUAAAGUUCAACAACAAACAGAGCCAUAAACAUUCCCAU_UACAAAUUAACACUUC______AUAUAUAUUC ..............(((((....)))))((((.....))))............................................................... ( -1.54 = -3.18 + 1.64)



| Location | 3,814,500 – 3,814,602 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -9.97 |
| Energy contribution | -12.63 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3814500 102 + 22407834 UGGGAAUGUUUAUGGCUCUGUUUGUUGUUGAACUUUAUGUUCCAACCAAUUGGUUG-GUU--UGUGGAGCAA-UGGGGGAAAUUCCCAGAGGGAGACAUUAAUUUA ...((((....(((.((((.(((.(..(((..(((((((..((((((....)))))-)..--))))))))))-..)(((.....)))))).)))).)))..)))). ( -30.90) >DroSec_CAF1 3515 101 + 1 UGGGAAUGUUUAUGGCUCUGUUUGUUGUGGAACUUUAUGUUCCAACCAAUUGGUUG-GUU--UGUGGAGCAA-UAGGGGA-AUUCCCAGAGGGAGACAAUAAUUUA (((((((.(((((.((((..(.......(((((.....)))))((((((....)))-)))--.)..)))).)-))))...-)))))))...(....)......... ( -31.70) >DroSim_CAF1 3505 101 + 1 UGGGAAUGUUUAUGGCUCUGUUUGUUGUGGAACUUUAUGUUCCAACCAAUUGGUUG-GUU--UGUGAAGCAA-UAGGGGA-AUUCCCAGACGGAGACCAUAAUUAA .........((((((((((((((.........(((((((..((((((....)))))-)..--)))))))...-...(((.-...))))))))))).)))))).... ( -36.70) >DroEre_CAF1 7774 94 + 1 UGGGAAUGUUUAUGGCUCUGUUUGUUGUGGAACUUUAUGUUCCAACCAAUUGGUUG-GUUCUUGUGGCGCAA-UGGGU----------GAGGGUGACAUUAAUUUA ....((((((....((((((..(((((((..((.....(..((((((....)))))-)..)..))..)))))-))..)----------.)))))))))))...... ( -25.80) >DroWil_CAF1 13372 85 + 1 UGAUUAUGAUGAUG---AUGUUUGUUGUUCAAUUUUAUGACUCUACAAAUUGGUUCAGCA--UGAAAAGCAAAUAUAGGA-AUUACCU--------------UUA- ......((((....---(((((((((.((((......(((..(........)..)))...--)))).)))))))))....-))))...--------------...- ( -13.00) >DroYak_CAF1 3448 102 + 1 UGGAAAUGUUUAUGGCACUGUUUGUUGUGGAACUUUAUGUUCCAACCAAUUGGUUG-GUU-UGGUGGAGCAA-UGGGGCA-AUUCCCAGAGGGUGACAUUAAUUUA ...((((....(((.((((.((((....((((((((((((((((.((((.......-..)-))))))))).)-)))))..-.)))))))).)))).)))..)))). ( -27.60) >consensus UGGGAAUGUUUAUGGCUCUGUUUGUUGUGGAACUUUAUGUUCCAACCAAUUGGUUG_GUU__UGUGGAGCAA_UAGGGGA_AUUCCCAGAGGGAGACAUUAAUUUA .((((((........(((((.(((((..(((((.....)))))((((....))))............))))).)))))...))))))................... ( -9.97 = -12.63 + 2.67)

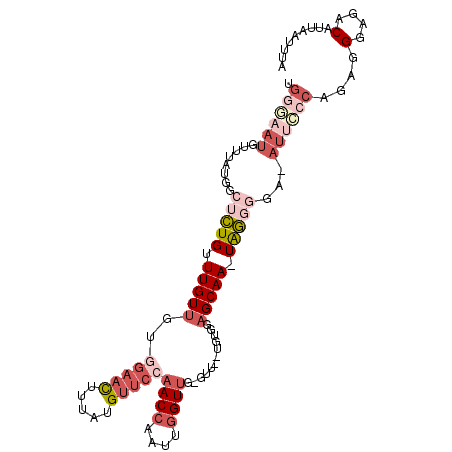
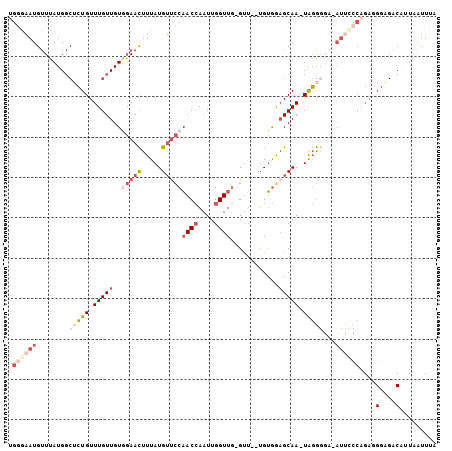
| Location | 3,814,500 – 3,814,602 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -7.75 |
| Energy contribution | -9.37 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3814500 102 - 22407834 UAAAUUAAUGUCUCCCUCUGGGAAUUUCCCCCA-UUGCUCCACA--AAC-CAACCAAUUGGUUGGAACAUAAAGUUCAACAACAAACAGAGCCAUAAACAUUCCCA ...................((((((........-..((((....--...-(((((....)))))((((.....))))...........)))).......)))))). ( -21.13) >DroSec_CAF1 3515 101 - 1 UAAAUUAUUGUCUCCCUCUGGGAAU-UCCCCUA-UUGCUCCACA--AAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCA ...................((((((-.....((-(.((((....--(((-(((....))))))(((((.....)))))..........)))).)))...)))))). ( -24.40) >DroSim_CAF1 3505 101 - 1 UUAAUUAUGGUCUCCGUCUGGGAAU-UCCCCUA-UUGCUUCACA--AAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCA ....((((((.(((.((..(((...-.)))...-(((.......--(((-(((....))))))(((((.....)))))....))))).)))))))))......... ( -25.10) >DroEre_CAF1 7774 94 - 1 UAAAUUAAUGUCACCCUC----------ACCCA-UUGCGCCACAAGAAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCA ......(((((....(((----------.....-............(((-(((....))))))(((((.....)))))..........)))......))))).... ( -14.60) >DroWil_CAF1 13372 85 - 1 -UAA--------------AGGUAAU-UCCUAUAUUUGCUUUUCA--UGCUGAACCAAUUUGUAGAGUCAUAAAAUUGAACAACAAACAU---CAUCAUCAUAAUCA -.((--------------((((((.-........)))))))).(--((.(((.....(((((....(((......)))...)))))...---..))).)))..... ( -9.90) >DroYak_CAF1 3448 102 - 1 UAAAUUAAUGUCACCCUCUGGGAAU-UGCCCCA-UUGCUCCACCA-AAC-CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGUGCCAUAAACAUUUCCA .......(((.(((....((((...-...))))-(((........-(((-(((....))))))(((((.....)))))....)))...))).)))........... ( -19.70) >consensus UAAAUUAAUGUCUCCCUCUGGGAAU_UCCCCCA_UUGCUCCACA__AAC_CAACCAAUUGGUUGGAACAUAAAGUUCCACAACAAACAGAGCCAUAAACAUUCCCA ...................((((((.........................(((((....)))))((((.....))))......................)))))). ( -7.75 = -9.37 + 1.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:31 2006