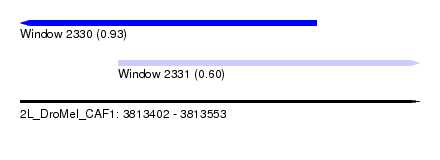
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,813,402 – 3,813,553 |
| Length | 151 |
| Max. P | 0.934940 |

| Location | 3,813,402 – 3,813,514 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
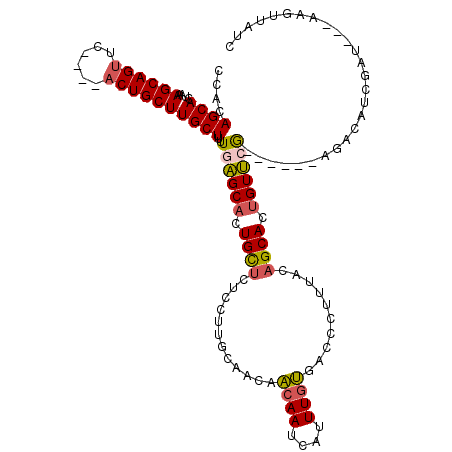
>2L_DroMel_CAF1 3813402 112 - 22407834 CCACAGCACCAAAGCAGUUU----ACUGCUUGCUUUUGAGCACUGCUCUCCUUGCAACAACAAUCAUUUGUGUCCCUUUACAGCACUGUUCGGCUCACAGACAUCGAU---AAGUUAUC .....(((...((((((...----.))))))......((((...))))....)))...........((((((..((...(((....)))..))..)))))).......---........ ( -23.10) >DroSec_CAF1 2417 107 - 1 CCACAGCAAUAAAGCAGUUC----ACUGCUUGCUUUUGAGCACUGCUCUCCUUGCAACAACAACCAUUUGUGGGCCUUUACAGCACCGUUCA-----CAGACAUCGAU---AAGUUAUC .....((((..((((((...----.))))))......((((...))))...))))...........(((((((((............)))))-----)))).......---........ ( -22.00) >DroSim_CAF1 2412 111 - 1 CCACAGCAGUAAAGCAGUUC----UCUGCUUGCUCUUGAGCACUGCUCUCCUUGCAACAACAAUCAUUUGUGGUCCUUUACAGCACUGUUCGGUAC-AAGACAUCGAU---AAGUUAUC ((((((((((.((((((...----.))))))(((....)))))))).....(((......))).....)))))...........(((..(((((..-.....))))).---.))).... ( -25.90) >DroEre_CAF1 2432 108 - 1 ACACAGCACUAAAGCAGUGCGCGCACUGCUUGCUUUUGCGCACUGUUAUCGGUUCAAUAGCAAUUGUUUUCGACCCUCCACUGCACUGUGAG--------CGAUAAAU---GUGUUAUC (((((((....((((((((....))))))))((....))))....(((((((((....(((....)))...))))...(((......)))..--------.))))).)---)))).... ( -30.30) >DroYak_CAF1 2339 111 - 1 CCACAGCACCUAAGCAGUGCACGAACUGCUUGCUUUUGCGCACUGUUAUCGCUUGAACAGCAAUAGCUUGCGACCCUUCACAGCACUGUGAA--------CGAUAAGUGCUGUGUUAUC .((((((((.((((((((......))))))))......((((..(((((.(((.....))).))))).))))..(.((((((....))))))--------.)....))))))))..... ( -40.10) >consensus CCACAGCACUAAAGCAGUUC____ACUGCUUGCUUUUGAGCACUGCUCUCCUUGCAACAACAAUCAUUUGUGACCCUUUACAGCACUGUUCG______AGACAUCGAU___AAGUUAUC ....((((....((((((......))))))))))..((((((.((((............((((....))))..........)))).))))))........................... (-17.41 = -17.17 + -0.24)

| Location | 3,813,439 – 3,813,553 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
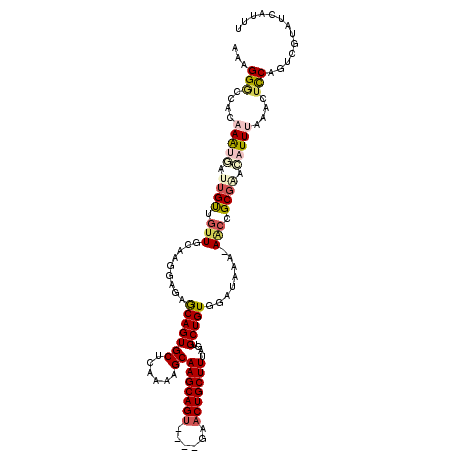
>2L_DroMel_CAF1 3813439 114 + 22407834 AAAGGGACACAAAUGAUUGUUGUUGCAAGGAGAGCAGUGCUCAAAAGCAAGCAGU----AAACUGCUUUGGUGCUGUGGCGGAA-ACCCGCGGAGAUUUAAACUCCUGUCGUAUCAUUU ...(.(((((((....))).)))).).(((((.((((..(.(((.....(((((.----...)))))))))..)))).((((..-..))))...........)))))............ ( -34.30) >DroSec_CAF1 2449 114 + 1 AAAGGCCCACAAAUGGUUGUUGUUGCAAGGAGAGCAGUGCUCAAAAGCAAGCAGU----GAACUGCUUUAUUGCUGUGGAUAAA-AACCGCGCACAUUUAAACUCCACUCGUAUCAUUU ..........((((((((((....))))(((((((((((((....)))((((((.----...)))))).))))))((((.....-..))))...........)))).......)))))) ( -30.20) >DroSim_CAF1 2448 115 + 1 AAAGGACCACAAAUGAUUGUUGUUGCAAGGAGAGCAGUGCUCAAGAGCAAGCAGA----GAACUGCUUUACUGCUGUGGAUAAAAAACCGCGAACAUUUAAACUUCAGUCGUAUCAUUU ............(((((((..(((.....(..(((((((((....)))((((((.----...)))))).))))))((((........))))...).....)))..)))))))....... ( -29.80) >DroEre_CAF1 2461 118 + 1 GGAGGGUCGAAAACAAUUGCUAUUGAACCGAUAACAGUGCGCAAAAGCAAGCAGUGCGCGCACUGCUUUAGUGCUGUGUAUAAA-AACCGCUAAUAUUCAAACUCCAUACUUAUCAGCU ((((.((((....((((....))))...)))).((((..(((....))((((((((....))))))))..)..)))).......-.................))))............. ( -32.10) >DroYak_CAF1 2371 117 + 1 GAAGGGUCGCAAGCUAUUGCUGUUCAAGCGAUAACAGUGCGCAAAAGCAAGCAGUUCGUGCACUGCUUAGGUGCUGUGGUUA-A-AGCCGCGCACAUUCAAACUCCAUUCUUAUCAGUU ((((((((((.(((....)))......)))))....((((((...((((((((((......))))))....))))..(((..-.-.))))))))).........)).)))......... ( -36.10) >consensus AAAGGGCCACAAAUGAUUGUUGUUGCAAGGAGAGCAGUGCUCAAAAGCAAGCAGU____GAACUGCUUUAGUGCUGUGGAUAAA_AACCGCGAACAUUUAAACUCCAGUCGUAUCAUUU ...(((....(((((.((((.(((.........(((((((......))(((((((......)))))))....)))))........))).)))).)))))....)))............. (-18.23 = -19.55 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:28 2006