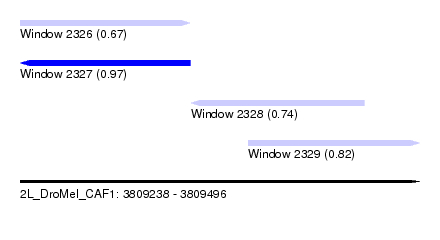
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,809,238 – 3,809,496 |
| Length | 258 |
| Max. P | 0.967697 |

| Location | 3,809,238 – 3,809,348 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -18.84 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
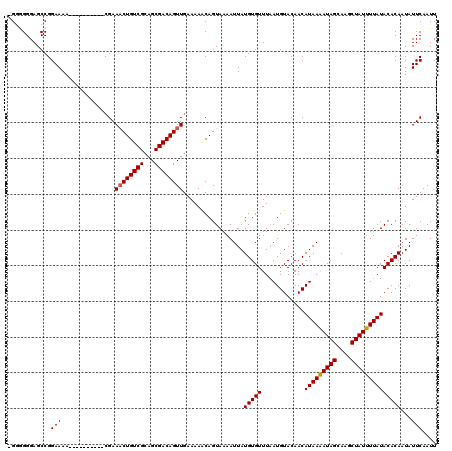
>2L_DroMel_CAF1 3809238 110 + 22407834 GGGGGGGAGCCGGAAAA----------CGAAACUGUCGCAGCGACAGUUGAAAAGCAGUAAAAUUAUGUGUUUAAUGUACAACAUAAAAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU ...((....)).(((..----------...((((((((...)))))))).................(((((............(((((((((....))))))))))))))...))).... ( -21.90) >DroSec_CAF1 18513 109 + 1 -GGGGGGAGCCGGAAAA----------CGAAUCUGUCGCAGCGACAGUUGAAAAACAGUAAAAUUAUGUGUUUUAUGUACAACAUAAGAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU -..((....))......----------.....((((((...))))))(((((..(((.((((((.....))))))))).....(((((((((....)))))))))........))))).. ( -20.80) >DroSim_CAF1 18513 109 + 1 -GGGGGGAGCCGGAAAA----------CGAAACUGUCGCAGCGACAGUUGAAAAACAGUAAAAUUAUGUGUUUAAUGUACAACAUAAAAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU -..((....)).(((..----------...((((((((...)))))))).................(((((............(((((((((....))))))))))))))...))).... ( -21.90) >DroEre_CAF1 20006 108 + 1 -G--GGGAGCCGGAAAA---------UCGAAACUGUCGCAGCGACAGUUUAAAAGCAGUAAAAUUAUGUGUUUAAUGUACAACAUAAAAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU -(--(....)).(((..---------...(((((((((...)))))))))................(((((............(((((((((....))))))))))))))...))).... ( -22.90) >DroYak_CAF1 20377 118 + 1 -G-AGGGAGCCGGAAAACGAAACGAAACGAAACUGUCGCAGCGACAGUUGAAAAUCAGUAAAAUUAUGUGUUUAAUGUACAACAUAAAAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU -.-...............(((..((.....((((((((...)))))))).....))..........(((((............(((((((((....))))))))))))))...))).... ( -21.00) >consensus _GGGGGGAGCCGGAAAA__________CGAAACUGUCGCAGCGACAGUUGAAAAACAGUAAAAUUAUGUGUUUAAUGUACAACAUAAAAUAGCAAGCUAUUUUAUACACAAUAUUCAAUU ............(((...............((((((((...)))))))).................(((((............(((((((((....))))))))))))))...))).... (-18.84 = -18.88 + 0.04)

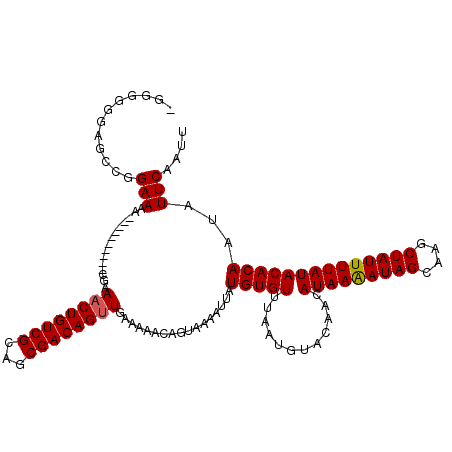
| Location | 3,809,238 – 3,809,348 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3809238 110 - 22407834 AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUUUUAUGUUGUACAUUAAACACAUAAUUUUACUGCUUUUCAACUGUCGCUGCGACAGUUUCG----------UUUUCCGGCUCCCCCCC ...((((.((((((((((((((((....)))))))))(((........))))))))))))))..((.....((((((((...)))))))).((----------.....))))........ ( -21.40) >DroSec_CAF1 18513 109 - 1 AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUCUUAUGUUGUACAUAAAACACAUAAUUUUACUGUUUUUCAACUGUCGCUGCGACAGAUUCG----------UUUUCCGGCUCCCCCC- ....((((.((((((((((.((((....))))......)))))))))).........................((((((...)))))))))).----------................- ( -18.30) >DroSim_CAF1 18513 109 - 1 AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUUUUAUGUUGUACAUUAAACACAUAAUUUUACUGUUUUUCAACUGUCGCUGCGACAGUUUCG----------UUUUCCGGCUCCCCCC- ...((((.((((((((((((((((....)))))))))(((........)))))))))))))).........((((((((...))))))))...----------................- ( -21.00) >DroEre_CAF1 20006 108 - 1 AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUUUUAUGUUGUACAUUAAACACAUAAUUUUACUGCUUUUAAACUGUCGCUGCGACAGUUUCGA---------UUUUCCGGCUCCC--C- ((((((..((((((((((((((((....)))))))))(((........)))))))))).............((((((((...))))))))))))---------))...........--.- ( -22.90) >DroYak_CAF1 20377 118 - 1 AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUUUUAUGUUGUACAUUAAACACAUAAUUUUACUGAUUUUCAACUGUCGCUGCGACAGUUUCGUUUCGUUUCGUUUUCCGGCUCCCU-C- ...((((.((((((((((((((((....)))))))))(((........))))))))))))))..((.....((((((((...)))))))).....))....................-.- ( -22.00) >consensus AAUUGAAUAUUGUGUAUAAAAUAGCUUGCUAUUUUAUGUUGUACAUUAAACACAUAAUUUUACUGCUUUUCAACUGUCGCUGCGACAGUUUCG__________UUUUCCGGCUCCCCCC_ ...((((.((((((((((((((((....)))))))))(((........)))))))))))))).........((((((((...)))))))).............................. (-19.62 = -20.02 + 0.40)

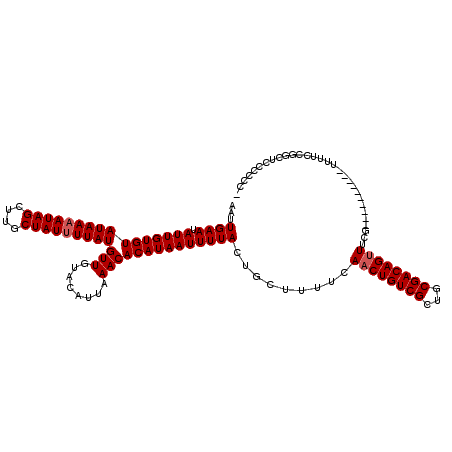
| Location | 3,809,348 – 3,809,460 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3809348 112 - 22407834 GUUGUCCACCAAGCAAUCAUCAAUCAU-AUUGUUUAAACUAAAGAGCUUUAAUCGUAUG----CAUGAGUGUUGAUGGCAGCUUUUUGGUUU---AAAUCGUUUCUCUUAGAUGUGGAUA ..(((((((..................-...(((((((((((((((((...((((.(((----(....))))))))...)))))))))))))---))))....((.....)).))))))) ( -29.40) >DroSec_CAF1 18622 112 - 1 GUUGUCCACCAAGCAAUCAUCAAUCAU-AUUGGUAAAACUAAAGAGCUUUAAUCGUAUG----CAUGAGUGUUGAUGGCACCUUUUUGGUUU---AAAUCGUUUCUCUUAGAUGUGGAUA ..(((((((.........((((((...-)))))).(((((((((((.....((((.(((----(....)))))))).....)))))))))))---..................))))))) ( -22.80) >DroSim_CAF1 18622 112 - 1 GUUGUCCACCAAGCAAUCAUCAAUCAU-AUUGGUAAAACUAAAGAGCUUUAAUCGUAUG----CAUGAGUGUUGAUGGCAGCUUUUUGGUUU---AAAUCGUUUCUCUUAGAUGUGGAUA ..(((((((.........((((((...-)))))).(((((((((((((...((((.(((----(....))))))))...)))))))))))))---..................))))))) ( -28.10) >DroEre_CAF1 20114 116 - 1 GUUGUCCACCAAGCAAUCAUCAAUCAA-AUUGGUGAAACUAAAGAGCUUUAAUCGUAUGCAUGCAUGAGUGUUGAUGGCAGCUUUUUGGUUU---UAAUCGUUUCUCUUAAAUGUGGAUA ..(((((((.......((((((((...-))))))))((((((((((((...((((...((((......))))))))...)))))))))))).---..................))))))) ( -31.40) >DroYak_CAF1 20495 116 - 1 GCUGUCCACCAAGCAAUCAACAAUCAAUAUUGGUGAAACUAAAGAGCUUUAAUCGUAUG----CAUGAGUGUUGAUGGCAGCUUUUUGGUUUCGUUAAUCGUUUCUCUUAAAUGUGGAUA ..(((((((...................(((..(((((((((((((((...((((.(((----(....))))))))...)))))))))))))))..))).((((.....))))))))))) ( -31.10) >consensus GUUGUCCACCAAGCAAUCAUCAAUCAU_AUUGGUAAAACUAAAGAGCUUUAAUCGUAUG____CAUGAGUGUUGAUGGCAGCUUUUUGGUUU___AAAUCGUUUCUCUUAGAUGUGGAUA ..(((((((.........((((((....)))))).(((((((((((((...((((((((....)))).....))))...))))))))))))).....................))))))) (-22.12 = -22.72 + 0.60)



| Location | 3,809,385 – 3,809,496 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3809385 111 + 22407834 UGCCAUCAACACUCAUG----CAUACGAUUAAAGCUCUUUAGUUUAAACAAU-AUGAUUGAUGAUUGCUUGGUGGACAACUAGUAAGCGCAACAACUAAAC---AAUUGGCAAAUGUU-U ((((((((.((.(((((----.....(((((((....))))))).......)-)))).)).)))..(((((.(((....))).))))).............---...)))))......-. ( -22.70) >DroSec_CAF1 18659 111 + 1 UGCCAUCAACACUCAUG----CAUACGAUUAAAGCUCUUUAGUUUUACCAAU-AUGAUUGAUGAUUGCUUGGUGGACAACUAGUAAGCGCAACAACUAAAC---AAUUGGCAAAUGUU-U ((((((((.((.(((((----..((.(((((((....))))))).))....)-)))).)).)))..(((((.(((....))).))))).............---...)))))......-. ( -23.00) >DroSim_CAF1 18659 111 + 1 UGCCAUCAACACUCAUG----CAUACGAUUAAAGCUCUUUAGUUUUACCAAU-AUGAUUGAUGAUUGCUUGGUGGACAACUAGUAAGCGCAACAACUAAAC---AAUUGGCAAAUGUU-U ((((((((.((.(((((----..((.(((((((....))))))).))....)-)))).)).)))..(((((.(((....))).))))).............---...)))))......-. ( -23.00) >DroEre_CAF1 20151 116 + 1 UGCCAUCAACACUCAUGCAUGCAUACGAUUAAAGCUCUUUAGUUUCACCAAU-UUGAUUGAUGAUUGCUUGGUGGACAACUAGUAAGCGCAACAACUAAAC---AAUUGGCAAAUGUUUU (((((.........(((....))).........(((..(((((((((((((.-..((((...))))..))))))))..)))))..))).............---...)))))........ ( -23.20) >DroYak_CAF1 20535 115 + 1 UGCCAUCAACACUCAUG----CAUACGAUUAAAGCUCUUUAGUUUCACCAAUAUUGAUUGUUGAUUGCUUGGUGGACAGCUAUUAAGCGCAACAACUAAACAACAAUUGGCAAAUGUU-U (((((((((((.(((..----.....(((((((....)))))))..........))).))))))..(((((((((....)))))))))...................)))))......-. ( -26.33) >consensus UGCCAUCAACACUCAUG____CAUACGAUUAAAGCUCUUUAGUUUCACCAAU_AUGAUUGAUGAUUGCUUGGUGGACAACUAGUAAGCGCAACAACUAAAC___AAUUGGCAAAUGUU_U ((((((((.((.((((..........(((((((....))))))).........)))).)).)))..(((((.(((....))).)))))...................)))))........ (-21.05 = -21.45 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:26 2006