
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,807,724 – 3,807,993 |
| Length | 269 |
| Max. P | 0.999294 |

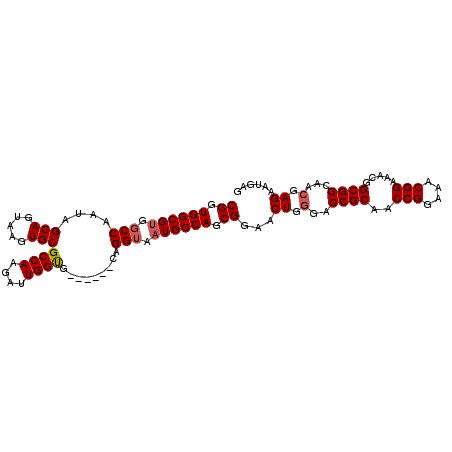
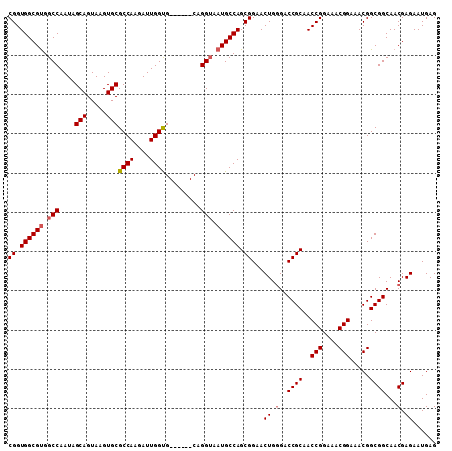
| Location | 3,807,724 – 3,807,833 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -35.82 |
| Energy contribution | -36.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3807724 109 + 22407834 CGGUGGCGCGGCCAAUAGCAGUAAGUGCGCCAAGAUUGGCGCACUGGCAGGGAAUGCCAGCGGAACUGUGGCCGCAACCGGAAACGGAAACGGCGGCAACGAGAAUGAG ..((.((((((((....(((((.(((((((((....)))))))))((((.....))))......)))))))))))..(((....))).....)).))............ ( -53.50) >DroSec_CAF1 17032 103 + 1 CGGUGGCGUGGCCAAUAGCAGUAAGUGCGCCAAGAUUGGUG------CAGGUAAUGCCAGCGGAACUGGGACCGCAACCGGAAACGGAAACGGCGGCAACGAGAAUGAG ((.((((((.(((....((.....))((((((....)))))------).))).)))))).))...((.(..((((..(((....))).....))))...).))...... ( -41.20) >DroSim_CAF1 17023 103 + 1 CGGUGGCGUGGCCAAUAGCAGUAAGUGCGCCAAGAUUGGUG------CAGGUAAUGCCAGCGGAACUGGGACCGCAACCGGAAACGGAAACGGCGGCAACGAGAAUGAG ((.((((((.(((....((.....))((((((....)))))------).))).)))))).))...((.(..((((..(((....))).....))))...).))...... ( -41.20) >consensus CGGUGGCGUGGCCAAUAGCAGUAAGUGCGCCAAGAUUGGUG______CAGGUAAUGCCAGCGGAACUGGGACCGCAACCGGAAACGGAAACGGCGGCAACGAGAAUGAG ((.((((((.(((....(((.....)))((((....)))).........))).)))))).))...((.(..((((..(((....))).....))))...).))...... (-35.82 = -36.60 + 0.78)

| Location | 3,807,724 – 3,807,833 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -27.53 |
| Energy contribution | -27.03 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3807724 109 - 22407834 CUCAUUCUCGUUGCCGCCGUUUCCGUUUCCGGUUGCGGCCACAGUUCCGCUGGCAUUCCCUGCCAGUGCGCCAAUCUUGGCGCACUUACUGCUAUUGGCCGCGCCACCG ........(((.((((((((..(((....)))..)))))..((((...(((((((.....)))))))((((((....))))))....)))).....))).)))...... ( -45.80) >DroSec_CAF1 17032 103 - 1 CUCAUUCUCGUUGCCGCCGUUUCCGUUUCCGGUUGCGGUCCCAGUUCCGCUGGCAUUACCUG------CACCAAUCUUGGCGCACUUACUGCUAUUGGCCACGCCACCG ........(((.((((((((..(((....)))..)))))...........(((((.((..((------(.(((....))).)))..)).)))))..))).)))...... ( -31.40) >DroSim_CAF1 17023 103 - 1 CUCAUUCUCGUUGCCGCCGUUUCCGUUUCCGGUUGCGGUCCCAGUUCCGCUGGCAUUACCUG------CACCAAUCUUGGCGCACUUACUGCUAUUGGCCACGCCACCG ........(((.((((((((..(((....)))..)))))...........(((((.((..((------(.(((....))).)))..)).)))))..))).)))...... ( -31.40) >consensus CUCAUUCUCGUUGCCGCCGUUUCCGUUUCCGGUUGCGGUCCCAGUUCCGCUGGCAUUACCUG______CACCAAUCUUGGCGCACUUACUGCUAUUGGCCACGCCACCG ........(((.((((((((..(((....)))..)))))..((((..(((..(.(((...(((....)))..))).)..))).....)))).....))).)))...... (-27.53 = -27.03 + -0.49)

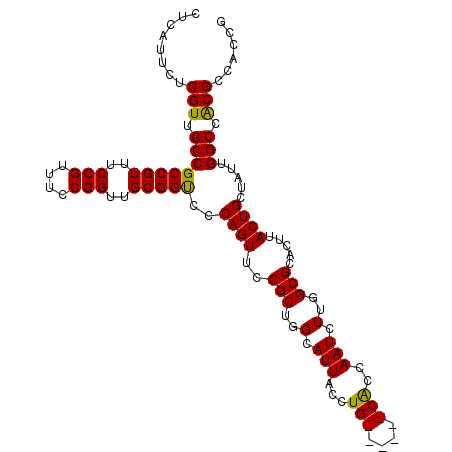
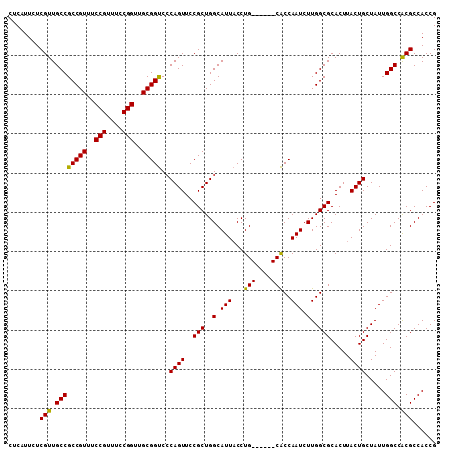
| Location | 3,807,759 – 3,807,873 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -29.83 |
| Energy contribution | -29.31 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3807759 114 + 22407834 UUGGCGCACUGGCAGGGAAUGCCAGCGGAACUGUGGCCGCAACCG------GAAACGGAAACGGCGGCAACGAGAAUGAGCCGAGUGUGAUCAAGGCGUGCAGCAGCUCGCUAAAGAUGA (((((((((((((.......(((.((((........))))..(((------....)))....)))(....)........))).)))))).))))((((.((....)).))))........ ( -45.00) >DroSec_CAF1 17067 108 + 1 UUGGUG------CAGGUAAUGCCAGCGGAACUGGGACCGCAACCG------GAAACGGAAACGGCGGCAACGAGAAUGAGCCGAGUGUGAUCAAGGCGUGCAGCAGCUCGCUGAAGAUGA (((((.------((......(((.((((........))))..(((------....)))....)))(....).....)).))))).....(((..((((.((....)).))))...))).. ( -36.20) >DroSim_CAF1 17058 108 + 1 UUGGUG------CAGGUAAUGCCAGCGGAACUGGGACCGCAACCG------GAAACGGAAACGGCGGCAACGAGAAUGAGCCGAGUGUGAUCAAGGCGUGCAGCAGCUCGCUGAAGAUGA (((((.------((......(((.((((........))))..(((------....)))....)))(....).....)).))))).....(((..((((.((....)).))))...))).. ( -36.20) >DroEre_CAF1 18535 108 + 1 UUGGCG------CAGGGAUUGCCAGCGGAACCGGAGCCGCAACCG------GAAACGGAAACGGUGGCAACGAGAACGAGCCGAGUGUGAUCAAGGCGUGCAGCAGCUCGCUGAAGAUGA ...(((------(...(((..(...(((..(((..(((((..(((------....))).....)))))..)).)......)))...)..)))...)))).((((.....))))....... ( -38.60) >DroYak_CAF1 18903 114 + 1 UUGGUG------AAGGGAAUGCCAGCGGAACGGGAGUGGCAACCGGCAACGGAAACGGAAACGGUGGCAACGAGAAUGAACCAAGUGUGAUCAAGGCGUGCAGCAGCUCGCUGAAGAUGA (((((.------.......(((((((...((....)).))..(((....)))...((....)).)))))..........))))).....(((..((((.((....)).))))...))).. ( -34.37) >consensus UUGGUG______CAGGGAAUGCCAGCGGAACUGGGGCCGCAACCG______GAAACGGAAACGGCGGCAACGAGAAUGAGCCGAGUGUGAUCAAGGCGUGCAGCAGCUCGCUGAAGAUGA (((((.........(....((((.((((........))))...............((....))..)))).)........))))).....(((..((((.((....)).))))...))).. (-29.83 = -29.31 + -0.52)

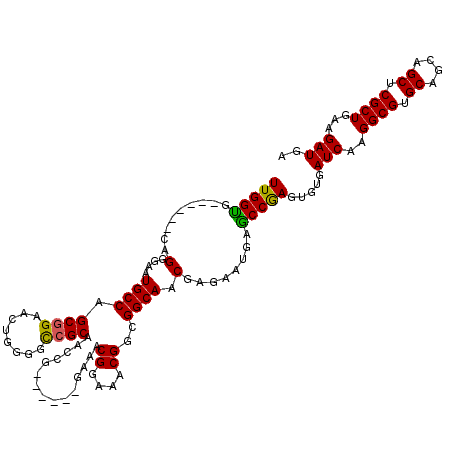
| Location | 3,807,833 – 3,807,953 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -49.40 |
| Consensus MFE | -49.14 |
| Energy contribution | -49.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3807833 120 - 22407834 GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUAUUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGCUCAUCUUUAGCGAGCUGCUGCACGCCUUGAUCACACUCGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..))...((((((((((((.((.((((.........)))).)).))).)))...))))))..... ( -49.70) >DroSec_CAF1 17135 120 - 1 GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGCUCAUCUUCAGCGAGCUGCUGCACGCCUUGAUCACACUCGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..))...((((((((((((.((.((((.........)))).)).))).)))...))))))..... ( -49.00) >DroSim_CAF1 17126 120 - 1 GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGAAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGCUCAUCUUCAGCGAGCUGCUGCACGCCUUGAUCACACUCGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..))...((((((((((((.((.((((.........)))).)).))).)))...))))))..... ( -49.00) >DroEre_CAF1 18603 120 - 1 GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAGAUGUGGUGGCUGCUGUUGCUCAUCUUCAGCGAGCUGCUGCACGCCUUGAUCACACUCGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..)).((((((((((((((.((.((((.........)))).)).))).)))...)))))).)).. ( -50.30) >DroYak_CAF1 18977 120 - 1 GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGCUCAUCUUCAGCGAGCUGCUGCACGCCUUGAUCACACUUGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..))...((((((((((((.((.((((.........)))).)).))).)))...))))))..... ( -49.00) >consensus GCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGCUCAUCUUCAGCGAGCUGCUGCACGCCUUGAUCACACUCGG ((..((((((....)))((.(((((.((((((....))))))...))))))))))..))...((((((((((((.((.((((.........)))).)).))).)))...))))))..... (-49.14 = -49.14 + 0.00)



| Location | 3,807,873 – 3,807,993 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -53.80 |
| Consensus MFE | -53.14 |
| Energy contribution | -53.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3807873 120 - 22407834 GCGGACUGACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUAUUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....))))))))).((((((((.(((((.((((((....))))))...)))))))))..))))..))))).)).))).))).. ( -53.70) >DroSec_CAF1 17175 120 - 1 GCGGACUGACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....))))))))).((((((((.(((((.((((((....))))))...)))))))))..))))..))))).)).))).))).. ( -53.00) >DroSim_CAF1 17166 120 - 1 GCGGACUGACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGAAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....))))))))).((((((((.(((((.((((((....))))))...)))))))))..))))..))))).)).))).))).. ( -53.00) >DroEre_CAF1 18643 120 - 1 GCGGACUGACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAGAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....)))))))))(((((((((.(((((.((((((....))))))...)))))))))..))))).))))).)).))).))).. ( -56.30) >DroYak_CAF1 19017 120 - 1 GCGGACUAACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....))))))))).((((((((.(((((.((((((....))))))...)))))))))..))))..))))).)).))).))).. ( -53.00) >consensus GCGGACUGACCGAACGCACCUCCAUAUCCAGGCCAGUGGUGCUGGCCGGACUGCUCCGCGAGCUGUUGUAGUGGAAACUAUACUUCGGCUGCGGCUGGUAAAUGUGGUGGCUGCUGUUGC .(((.....)))((((((((.((((((((.((((((.....))))))))).((((((((.(((((.((((((....))))))...)))))))))..))))..))))).)).))).))).. (-53.14 = -53.14 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:20 2006