
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
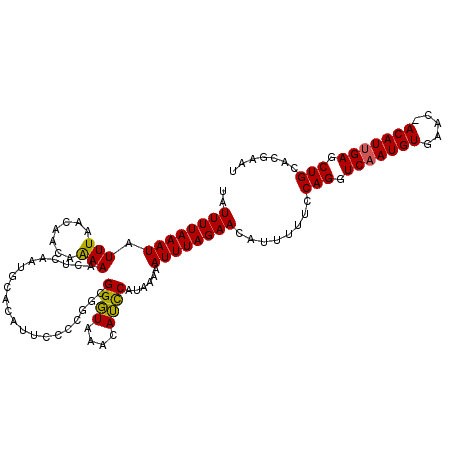
| Location | 3,744,713 – 3,744,825 |
| Length | 112 |
| Max. P | 0.601986 |

| Location | 3,744,713 – 3,744,825 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -13.40 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3744713 112 + 22407834 UAUUUUAAAUAUUUAACAACCAAAUCUCUAUUCACAUUUCCCGGCGGGUAAAUAUCCAUACAAAUUUAGAACAUUUUUCCAGGUCAAUGUGAAC-ACAUUGAGCUGCACGAAU ..((((((((.........((.....................)).((((....))))......))))))))........(((.(((((((....-))))))).)))....... ( -17.00) >DroSec_CAF1 206814 112 + 1 UAUUUUAAAUAUUUAACAACAGAAACUUAAUGCACAUUCCCCGGCGGGUAAACAUUCAUAAAAAUUUAGAACAUUUUUCCAGGUCAAUGUGAAC-ACAUUGAGCUGCACGAAU ..((((((((.((((.(....)......((((......(((....)))....))))..)))).))))))))........(((.(((((((....-))))))).)))....... ( -17.50) >DroSim_CAF1 213953 107 + 1 UAUUUUAAAUAUUUAACAGCAGAAACUCAAUGCACAUUCC-----GGGUAAACAUUCAUAAAAAUUUAGAACAUUUUUCCAGGUCAAUGUGAAC-ACAUUGAGCUGCACGAAU ..................((((...(((((((((((((((-----(((.(((..(((.(((....))))))..))).))).))..))))))...-.)))))))))))...... ( -21.50) >DroYak_CAF1 224759 113 + 1 UAUUUUAAAUAUUUAACAGCCAAAUCUCUAUACACAUUCCCCAUUGGCUAAACAGCCAUAAAAAUUUAGAACAUUUUUCCAGGUCAAUGUGAACGACAUGGAGCUGCACAAAU ................((((...((.((....((((((..((..(((((....)))))..........(((.....)))..))..))))))...)).))...))))....... ( -18.90) >consensus UAUUUUAAAUAUUUAACAACAAAAACUCAAUGCACAUUCCCCGGCGGGUAAACAUCCAUAAAAAUUUAGAACAUUUUUCCAGGUCAAUGUGAAC_ACAUUGAGCUGCACGAAU ..((((((((.(((.......))).....................((((....))))......))))))))........(((.(((((((.....))))))).)))....... (-13.40 = -12.77 + -0.62)

| Location | 3,744,713 – 3,744,825 |
|---|---|
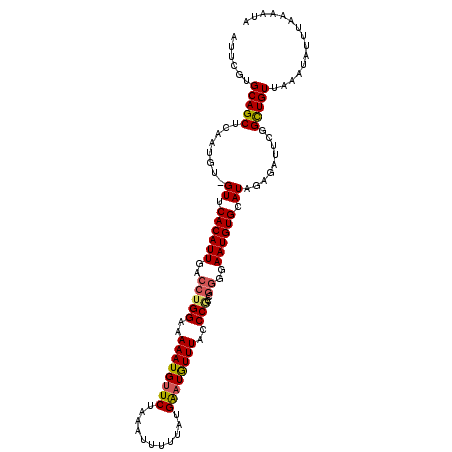
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3744713 112 - 22407834 AUUCGUGCAGCUCAAUGU-GUUCACAUUGACCUGGAAAAAUGUUCUAAAUUUGUAUGGAUAUUUACCCGCCGGGAAAUGUGAAUAGAGAUUUGGUUGUUAAAUAUUUAAAAUA ......((((((...(.(-(((((((((..(((((..((((((((...........)))))))).....))))).)))))))))).).....))))))............... ( -29.00) >DroSec_CAF1 206814 112 - 1 AUUCGUGCAGCUCAAUGU-GUUCACAUUGACCUGGAAAAAUGUUCUAAAUUUUUAUGAAUGUUUACCCGCCGGGGAAUGUGCAUUAAGUUUCUGUUGUUAAAUAUUUAAAAUA ....(..((..(((((((-....)))))))(((((..((((((((...........)))))))).....)))))...))..)............................... ( -23.10) >DroSim_CAF1 213953 107 - 1 AUUCGUGCAGCUCAAUGU-GUUCACAUUGACCUGGAAAAAUGUUCUAAAUUUUUAUGAAUGUUUACCC-----GGAAUGUGCAUUGAGUUUCUGCUGUUAAAUAUUUAAAAUA ......(((((((((((.-...((((((...((((..((((((((...........))))))))..))-----)))))))))))))))...)))).................. ( -25.40) >DroYak_CAF1 224759 113 - 1 AUUUGUGCAGCUCCAUGUCGUUCACAUUGACCUGGAAAAAUGUUCUAAAUUUUUAUGGCUGUUUAGCCAAUGGGGAAUGUGUAUAGAGAUUUGGCUGUUAAAUAUUUAAAAUA (((((.((((((..((.((...((((((..(((((((.....)))))........(((((....)))))..))..))))))....)).))..))))))))))).......... ( -29.90) >consensus AUUCGUGCAGCUCAAUGU_GUUCACAUUGACCUGGAAAAAUGUUCUAAAUUUUUAUGAAUGUUUACCCGCCGGGGAAUGUGCAUAGAGAUUCGGCUGUUAAAUAUUUAAAAUA ......(((((........((.((((((..(((((..((((((((...........))))))))..)))..))..)))))).)).........)))))............... (-17.82 = -18.00 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:49 2006