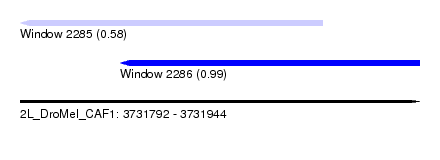
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,731,792 – 3,731,944 |
| Length | 152 |
| Max. P | 0.989167 |

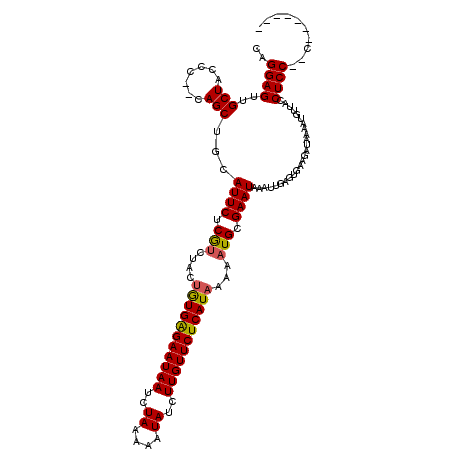
| Location | 3,731,792 – 3,731,907 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3731792 115 - 22407834 CAGGAGUUGCUACC---CAGCUGCAUUCCCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAAUAAAUUGAGUGAAGAUAAAAUGUUAGCUCCACC--GAGAG ..((((((((....---...((.(((((.(((....(((((((((((..((....))..))))))))))).....)))........))))).)).......).)))))))...--..... ( -23.54) >DroSec_CAF1 194047 108 - 1 CAGGAGUUGCUACCC--CAGCUGCAUUCCCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAAUAAAUUGAGUGAAGAUAAAAUGUUACCUCC---------- ..((((..(((....--.))).(((((...((((..(((((((((((..((....))..)))))))))))......(((.....))).....))))..)))))...))))---------- ( -18.70) >DroSim_CAF1 201200 108 - 1 CAGGAGUUGCUACCC--CAGCUGCAUUCUCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAAUAAAUUGAGUGAAGAUAAAAUGUUACCUCC---------- ..((((..((.....--...((.(((((((((....(((((((((((..((....))..))))))))))).....)))).......))))).)).......))...))))---------- ( -21.37) >DroEre_CAF1 207998 113 - 1 CAGGAGUUGCUACCCAGCAGCUGCAUUCUCAUCUGCCAUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUCAAAAUGCGAAUAAAUUGAGGGAAGAUAAAGUGUUACCUCCUUC------- ..(.(((((((....))))))).)((((.(((.(...((((((((((..((....))..))))))))))..).))).)))).....((((((.(.(((....)))).))))))------- ( -27.30) >DroYak_CAF1 207162 118 - 1 CAGGAGUUGCUAACC--CAGCUGCAUUCUCAGCUACUAUGGGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAAUAAAUUGUGUGAGGAUAAAAUGUUACCUCCACCUCCACAG ..((((((((.....--.(((((......)))))..(((((((((((..((....))..))))))))))).....)))).......(((.((((.(((....)))))))))))))).... ( -31.10) >consensus CAGGAGUUGCUACCC__CAGCUGCAUUCUCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAAUAAAUUGAGUGAAGAUAAAAUGUUACCUCC__C_______ ..((((..(((.......)))...((((.(((....(((((((((((..((....))..)))))))))))...))).)))).........................)))).......... (-18.76 = -18.52 + -0.24)

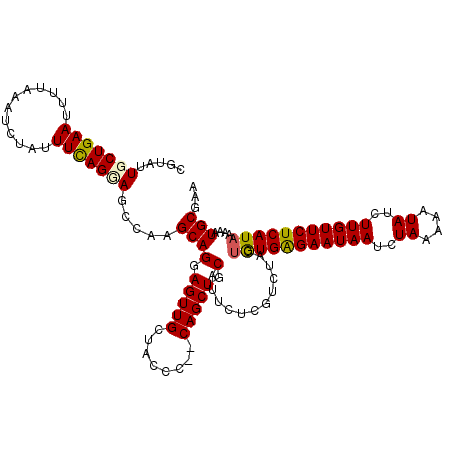
| Location | 3,731,830 – 3,731,944 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.49 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -22.72 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3731830 114 - 22407834 CAUAUUGCUGAAUUUUAAAUCUAUUUCAGUAGCCAAGCAGGAGUUGCUACC---CAGCUGCAUUCCCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAA ....((((((((............))))))))....((((((((.(((...---.)))...))))).......(((((((((((..((....))..)))))))))))....)))... ( -25.70) >DroSec_CAF1 194077 115 - 1 CGUAUUGCUGAAUUUUAAAUCUAUUUCAGGAGCCAAGCAGGAGUUGCUACCC--CAGCUGCAUUCCCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAA ((((((.(((((............))))).......((.(((((.(((....--.)))...))))).))....(((((((((((..((....))..)))))))))))..)))))).. ( -24.90) >DroSim_CAF1 201230 115 - 1 CGUAUUGCUGAAUUUUAAAUCUAUUUUAGUAGCCAAGCAGGAGUUGCUACCC--CAGCUGCAUUCUCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAA ((((((((((.....((((.....))))(((((..(((....))))))))..--))))...............(((((((((((..((....))..)))))))))))..)))))).. ( -25.10) >DroEre_CAF1 208031 117 - 1 CGUUGUGCUGAAUUUUAAAUCUAUUUCAGGAGCAAAGCAGGAGUUGCUACCCAGCAGCUGCAUUCUCAUCUGCCAUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUCAAAAUGCGAA ..((((.(((((............)))))..)))).((((.(((((((....))))))).).............((((((((((..((....))..)))))))))).....)))... ( -28.10) >DroYak_CAF1 207202 115 - 1 CGUGUUGCUGAAUUUUAAAUCUAUUUCAGCAGCAAAGCAGGAGUUGCUAACC--CAGCUGCAUUCUCAGCUACUAUGGGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAA ..((((((((((............))))))))))..(((((.((.....)))--)(((((......)))))..(((((((((((..((....))..)))))))))))....)))... ( -31.90) >consensus CGUAUUGCUGAAUUUUAAAUCUAUUUCAGGAGCCAAGCAGGAGUUGCUACCC__CAGCUGCAUUCUCGUCUACUGUGAGAAUAAUCUAAAAAUAUCUUGUUCUCAUAAAAAUGCGAA .....(((((((............))))))).....((((.(((((........))))).)............(((((((((((..((....))..)))))))))))....)))... (-22.72 = -22.68 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:45 2006