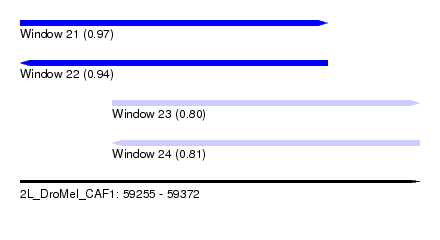
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 59,255 – 59,372 |
| Length | 117 |
| Max. P | 0.970469 |

| Location | 59,255 – 59,345 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -22.40 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
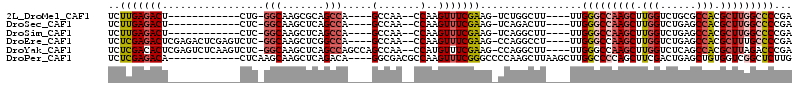
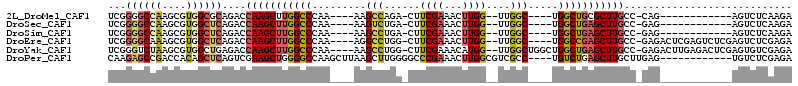
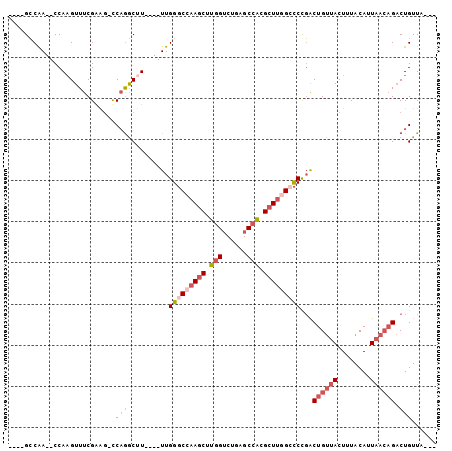
>2L_DroMel_CAF1 59255 90 + 22407834 UCUUGAGACU------------CUG-GGCAAGCGCAGCCA----GCCAA--CCAAGUUUCGAAG-UCUGGCUU----UUGGGCCAAGCUUGGUCUGCGCCACGCUUGGCCCCGA ..((((((((------------.((-(....((.......----))...--)))))))))))..-........----..(((((((((.((((....)))).)))))))))... ( -36.20) >DroSec_CAF1 32294 90 + 1 UCUUGAGACU------------CUC-GGCAAGCUCAGCCA----GCCAA--CCAAGUUUCGAAG-UCAGACUU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGA ..((((((((------------...-(((..((...))..----)))..--...))))))))..-........----..(((((((((.((((....)))).)))))))))... ( -34.90) >DroSim_CAF1 34610 90 + 1 UCUUGAGACU------------CUC-GGCAAGCUCAGCCA----GCCAA--CCAAGUUUCGAAG-UCAGGCUU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGA ..((((((((------------...-(((..((...))..----)))..--...))))))))..-........----..(((((((((.((((....)))).)))))))))... ( -34.90) >DroEre_CAF1 33727 102 + 1 UCUCGAGACUCGAGACUCGAGUCUC-GGCAAGCUCGGCCA----GCCAA--CCAAGUUUCGAAG-CCAGGCCU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUUGCCCCGA ....((((((((.....))))))))-(((((((..(((((----(...(--((((((((.(...-.).((((.----...)))))))))))))))).)))..)).))))).... ( -47.90) >DroYak_CAF1 32142 106 + 1 UCUCGACACUCGAGUCUCAAGUCUC-GGCAAGCUCAGCCAGCCAGCCAA--CCAUGUUUCGAAG-CCAGGCUU----UUGGGCCAAGCUUGGUCUCAGCCACGCUUAGACCCGA ..((((.((..(((.(....).)))-(((..(((.....)))..)))..--....)).))))((-(..((((.----...))))..))).(((((.(((...))).)))))... ( -32.40) >DroPer_CAF1 43093 98 + 1 UCUCGAGACA------------CUCAAGCAAGCUCAGACA----GGCGACGCCAAGUUUCGGGCCCCAAGCUUAAGCUUGGCCCCAGCUUCGACUGAGCUGUGGUCGGCUCUUG ....(((..(------------(.((....(((((((.((----(((...(((((((((.((((.....)))))))))))))....)))).).))))))).))))...)))... ( -36.00) >consensus UCUCGAGACU____________CUC_GGCAAGCUCAGCCA____GCCAA__CCAAGUUUCGAAG_CCAGGCUU____UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGA ..(((((((.................(((.......))).....(.......)..))))))).................(((((((((.(((......))).)))))))))... (-22.40 = -23.40 + 1.00)
| Location | 59,255 – 59,345 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 59255 90 - 22407834 UCGGGGCCAAGCGUGGCGCAGACCAAGCUUGGCCCAA----AAGCCAGA-CUUCGAAACUUGG--UUGGC----UGGCUGCGCUUGCC-CAG------------AGUCUCAAGA ((.((((.(((((..((((.(((((((..((((....----..))))(.-...)....)))))--)).))----..))..))))))))-).)------------)......... ( -38.20) >DroSec_CAF1 32294 90 - 1 UCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AAGUCUGA-CUUCGAAACUUGG--UUGGC----UGGCUGAGCUUGCC-GAG------------AGUCUCAAGA ...(((((((((.(((......))).)))))))))..----........-(((.((..(((((--(.(((----(.....)))).)))-)))------------..))..))). ( -38.50) >DroSim_CAF1 34610 90 - 1 UCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AAGCCUGA-CUUCGAAACUUGG--UUGGC----UGGCUGAGCUUGCC-GAG------------AGUCUCAAGA ...(((((((((.(((......))).)))))))))..----........-(((.((..(((((--(.(((----(.....)))).)))-)))------------..))..))). ( -38.50) >DroEre_CAF1 33727 102 - 1 UCGGGGCAAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AGGCCUGG-CUUCGAAACUUGG--UUGGC----UGGCCGAGCUUGCC-GAGACUCGAGUCUCGAGUCUCGAGA ((..(((((.((.((((.(((.(((((((.((((...----.)))).))-).((((...))))--)))))----)))))).)))))))-((((((((.....)))))))))).. ( -49.70) >DroYak_CAF1 32142 106 - 1 UCGGGUCUAAGCGUGGCUGAGACCAAGCUUGGCCCAA----AAGCCUGG-CUUCGAAACAUGG--UUGGCUGGCUGGCUGAGCUUGCC-GAGACUUGAGACUCGAGUGUCGAGA ((((((((.(((...))).))))((((((..(((...----.((((.((-((.........))--))))))....)))..))))))))-))..(((((.((....)).))))). ( -43.90) >DroPer_CAF1 43093 98 - 1 CAAGAGCCGACCACAGCUCAGUCGAAGCUGGGGCCAAGCUUAAGCUUGGGGCCCGAAACUUGGCGUCGCC----UGUCUGAGCUUGCUUGAG------------UGUCUCGAGA ...((((........))))..((((.(((.((((.(((((((..(..(((((((((...)))).))).))----.)..))))))))))).))------------)...)))).. ( -36.10) >consensus UCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA____AAGCCUGA_CUUCGAAACUUGG__UUGGC____UGGCUGAGCUUGCC_GAG____________AGUCUCAAGA ...((((((....))))))....(((((((((((.........(((......((((...))))....))).....)))))))))))............................ (-19.46 = -19.86 + 0.40)
| Location | 59,282 – 59,372 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -19.21 |
| Energy contribution | -21.10 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
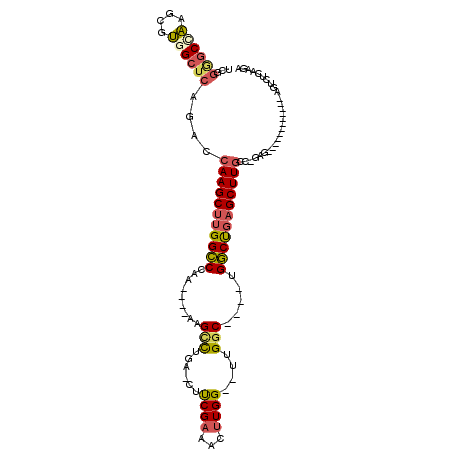
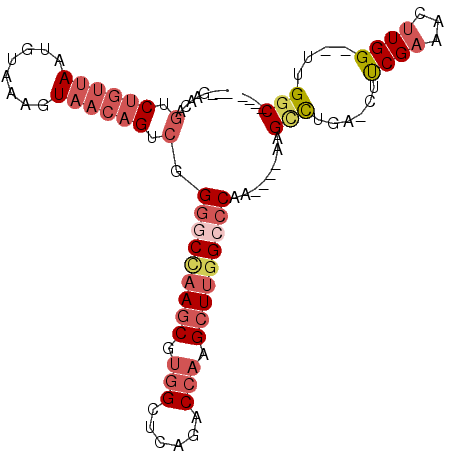
>2L_DroMel_CAF1 59282 90 + 22407834 ----GCCAA--CCAAGUUUCGAAG-UCUGGCUU----UUGGGCCAAGCUUGGUCUGCGCCACGCUUGGCCCCGACUGUUACUUUACAUUAACAGACUGUUC--- ----((((.--.(..........)-..))))..----..(((((((((.((((....)))).)))))))))...((((((........)))))).......--- ( -33.00) >DroSec_CAF1 32321 90 + 1 ----GCCAA--CCAAGUUUCGAAG-UCAGACUU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGACUGUUACUUUACAUUAACAGACUGUUC--- ----.....--.............-.(((....----..(((((((((.((((....)))).)))))))))...((((((........)))))).)))...--- ( -30.70) >DroSim_CAF1 34637 90 + 1 ----GCCAA--CCAAGUUUCGAAG-UCAGGCUU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGACUGUUACUUUACAUUAACAGACUGUUC--- ----(((..--.............-...)))..----..(((((((((.((((....)))).)))))))))...((((((........)))))).......--- ( -31.07) >DroEre_CAF1 33766 90 + 1 ----GCCAA--CCAAGUUUCGAAG-CCAGGCCU----UUGGGCCAAGCUUGGUCUGAGCCACGCUUUGCCCCGACUGUUACUUUACAUUAACAGACUGUUA--- ----.....--.............-.(((....----..((((.((((.((((....)))).)))).))))...((((((........)))))).)))...--- ( -24.60) >DroYak_CAF1 32181 97 + 1 GCCAGCCAA--CCAUGUUUCGAAG-CCAGGCUU----UUGGGCCAAGCUUGGUCUCAGCCACGCUUAGACCCGACUGUUACUUUACAAUAACAGACUGUUAUUG ..(((....--.......((((((-(..((((.----...))))..))))(((((.(((...))).))))))))((((((........)))))).)))...... ( -26.40) >DroPer_CAF1 43121 93 + 1 ----GGCGACGCCAAGUUUCGGGCCCCAAGCUUAAGCUUGGCCCCAGCUUCGACUGAGCUGUGGUCGGCUCUUGCUGUUACUUUACAGU----GCCUGUUA--- ----(((((.(((((((((.((((.....))))))))))((((.((((((.....)))))).)))))))))..(((((......)))))----))).....--- ( -36.00) >consensus ____GCCAA__CCAAGUUUCGAAG_CCAGGCUU____UUGGGCCAAGCUUGGUCUGAGCCACGCUUGGCCCCGACUGUUACUUUACAUUAACAGACUGUUA___ ..........................(((..........(((((((((.(((......))).)))))))))...((((((........)))))).)))...... (-19.21 = -21.10 + 1.89)

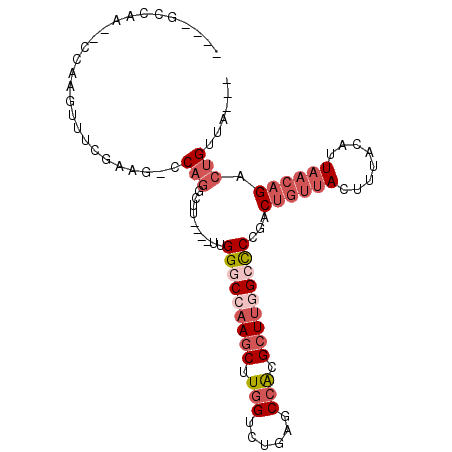
| Location | 59,282 – 59,372 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -19.50 |
| Energy contribution | -21.50 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 59282 90 - 22407834 ---GAACAGUCUGUUAAUGUAAAGUAACAGUCGGGGCCAAGCGUGGCGCAGACCAAGCUUGGCCCAA----AAGCCAGA-CUUCGAAACUUGG--UUGGC---- ---.....(.((((((........)))))).).(((((((((.(((......))).)))))))))..----..(((((.-((.........))--)))))---- ( -34.20) >DroSec_CAF1 32321 90 - 1 ---GAACAGUCUGUUAAUGUAAAGUAACAGUCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AAGUCUGA-CUUCGAAACUUGG--UUGGC---- ---.....(.((((((........)))))).).(((((((((.(((......))).)))))))))..----....(..(-((.........))--)..).---- ( -32.10) >DroSim_CAF1 34637 90 - 1 ---GAACAGUCUGUUAAUGUAAAGUAACAGUCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AAGCCUGA-CUUCGAAACUUGG--UUGGC---- ---.....(.((((((........)))))).).(((((((((.(((......))).)))))))))..----..(((.((-((.........))--)))))---- ( -33.90) >DroEre_CAF1 33766 90 - 1 ---UAACAGUCUGUUAAUGUAAAGUAACAGUCGGGGCAAAGCGUGGCUCAGACCAAGCUUGGCCCAA----AGGCCUGG-CUUCGAAACUUGG--UUGGC---- ---((((...((((((........))))))(((((((.((((.(((......))).))))((((...----.))))..)-))))))......)--)))..---- ( -28.90) >DroYak_CAF1 32181 97 - 1 CAAUAACAGUCUGUUAUUGUAAAGUAACAGUCGGGUCUAAGCGUGGCUGAGACCAAGCUUGGCCCAA----AAGCCUGG-CUUCGAAACAUGG--UUGGCUGGC (((((((.....)))))))........((((((((.((((((.(((......))).)))))))))..----.((((((.-........)).))--))))))).. ( -33.40) >DroPer_CAF1 43121 93 - 1 ---UAACAGGC----ACUGUAAAGUAACAGCAAGAGCCGACCACAGCUCAGUCGAAGCUGGGGCCAAGCUUAAGCUUGGGGCCCGAAACUUGGCGUCGCC---- ---.....(((----.((((......))))...(((((((...((((((....).)))))(((((.(((....)))...))))).....))))).)))))---- ( -34.50) >consensus ___GAACAGUCUGUUAAUGUAAAGUAACAGUCGGGGCCAAGCGUGGCUCAGACCAAGCUUGGCCCAA____AAGCCUGA_CUUCGAAACUUGG__UUGGC____ ........(.((((((........)))))).).(((((((((.(((......))).)))))))))........(((......((((...))))....))).... (-19.50 = -21.50 + 2.00)

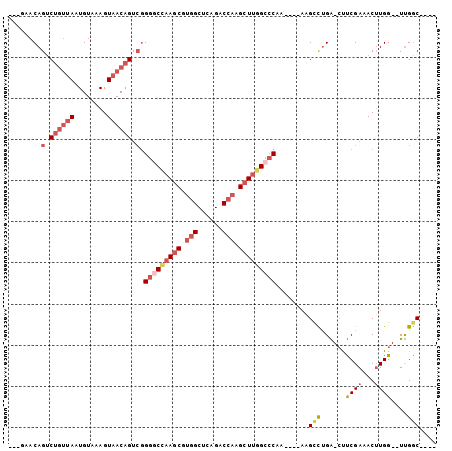
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:18 2006