
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,717,381 – 3,717,541 |
| Length | 160 |
| Max. P | 0.850565 |

| Location | 3,717,381 – 3,717,501 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -32.01 |
| Energy contribution | -31.79 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3717381 120 + 22407834 AUGGUCUCCAAAGAUUCAUGCAUGGCAAACUGCUCUCGCUCCAUCCUGUCCAGCUUCUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUCAAGGCAUCCAGCUGG ........(((((......((.(((((....((....)).......)).)))))........))))).((((((.((((((((...((((....))...))....)))))))))))))). ( -33.34) >DroSec_CAF1 179699 120 + 1 AUGGUCUUCAAAGAUUCCUGCAUGGCUAACUGCUCUCGCUCUAUCCUGUCCAGCUUAUCAUCCUUUGAUAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUUAAGGCAUCCAGCUGG .......((((((......((.((((.....((....))........).)))))........))))))((((((.((((((((...((((....))...))....)))))))))))))). ( -32.16) >DroSim_CAF1 188578 120 + 1 AUGGUCUUCAAAGACUCCUGCAUGGCUGACUGCUCUCGCUCUAUCCUGUCCAGCUUAUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAGACGUCUGCUUUUAAGGCAUCCAGCUGG .......((((((.....((...(((((((.((....))........)).)))))...))..))))))((((((.((((((((...((((....))...))....)))))))))))))). ( -35.90) >consensus AUGGUCUUCAAAGAUUCCUGCAUGGCUAACUGCUCUCGCUCUAUCCUGUCCAGCUUAUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUUAAGGCAUCCAGCUGG ........(((((......((.((((.....((....))........).)))))........))))).((((((.((((((((...((((....))...))....)))))))))))))). (-32.01 = -31.79 + -0.22)

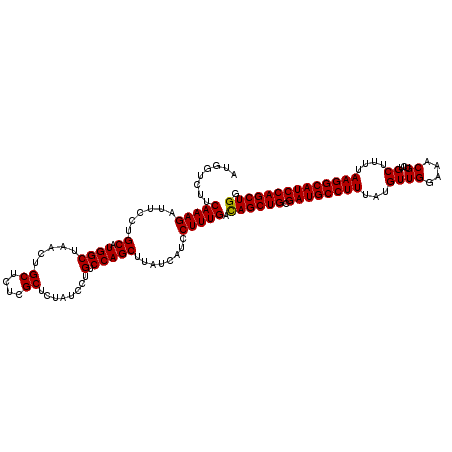
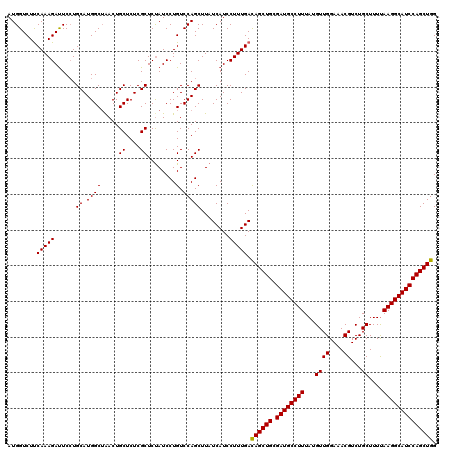
| Location | 3,717,381 – 3,717,501 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -35.19 |
| Consensus MFE | -33.90 |
| Energy contribution | -33.57 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3717381 120 - 22407834 CCAGCUGGAUGCCUUGAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAGAAGCUGGACAGGAUGGAGCGAGAGCAGUUUGCCAUGCAUGAAUCUUUGGAGACCAU .(((((((((((((((.((((....)))).))......))))))).))))))((.((((((.....(((((...((((...((....)).)))).))).))....)))))).))...... ( -37.30) >DroSec_CAF1 179699 120 - 1 CCAGCUGGAUGCCUUAAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUAUCAAAGGAUGAUAAGCUGGACAGGAUAGAGCGAGAGCAGUUAGCCAUGCAGGAAUCUUUGAAGACCAU ..(((((((((((((.......((....)).......)))))))).))))).((((((.((.....(((((....(((...((....)).)))..))).))....))))))))....... ( -31.54) >DroSim_CAF1 188578 120 - 1 CCAGCUGGAUGCCUUAAAAGCAGACGUCUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAUAAGCUGGACAGGAUAGAGCGAGAGCAGUCAGCCAUGCAGGAGUCUUUGAAGACCAU .((((((((((((((.......((....)).......)))))))).))))))(((((((.......(((((....(((...((....)).)))..))).)).....)))))))....... ( -36.74) >consensus CCAGCUGGAUGCCUUAAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAUAAGCUGGACAGGAUAGAGCGAGAGCAGUUAGCCAUGCAGGAAUCUUUGAAGACCAU .((((((((((((((.......((....)).......)))))))).))))))((((((.((.....(((((....(((...((....)).)))..))).))....))))))))....... (-33.90 = -33.57 + -0.33)


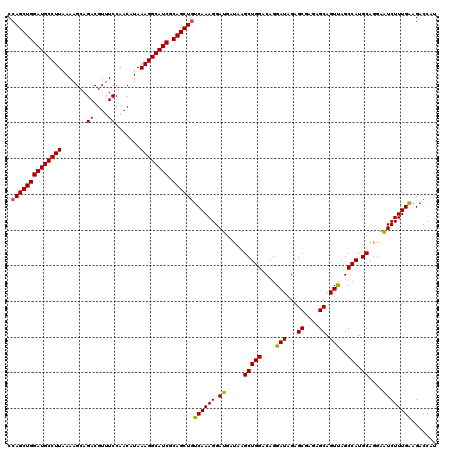
| Location | 3,717,421 – 3,717,541 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3717421 120 + 22407834 CCAUCCUGUCCAGCUUCUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUCAAGGCAUCCAGCUGGAUAUUCAACUGGAUCUGAUCCUUGCGGGCUUCGUUCGCUA ..(((..((((((....(((.....)))((((((.((((((((...((((....))...))....)))))))))))))).........))))))..)))....((((((...)))))).. ( -40.30) >DroSec_CAF1 179739 120 + 1 CUAUCCUGUCCAGCUUAUCAUCCUUUGAUAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUUAAGGCAUCCAGCUGGACACUCAACUGGUUCUGAUCCUUGACGGUUUCGUUCGCUA ......((((((((((((((.....)))))).((.((((((((...((((....))...))....))))))))))))))))))......((((..(((.((.....))..)))...)))) ( -34.80) >DroSim_CAF1 188618 120 + 1 CUAUCCUGUCCAGCUUAUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAGACGUCUGCUUUUAAGGCAUCCAGCUGGACACUCAACUGGUUCUGAUCCUUGAGGGUUUCGUUCGCUA ....(((((((......(((.....))).(((((.((((((((...((((....))...))....))))))))))))))))))(((((..((.......)))))))))............ ( -35.50) >consensus CUAUCCUGUCCAGCUUAUCAUCCUUUGACAGCUGCGAUGCCUUUAUGUUGGAAACGUCUGCUUUUAAGGCAUCCAGCUGGACACUCAACUGGUUCUGAUCCUUGAGGGUUUCGUUCGCUA .........((((....(((.....)))((((((.((((((((...((((....))...))....)))))))))))))).........))))....((.((.....))..))........ (-31.49 = -31.27 + -0.22)


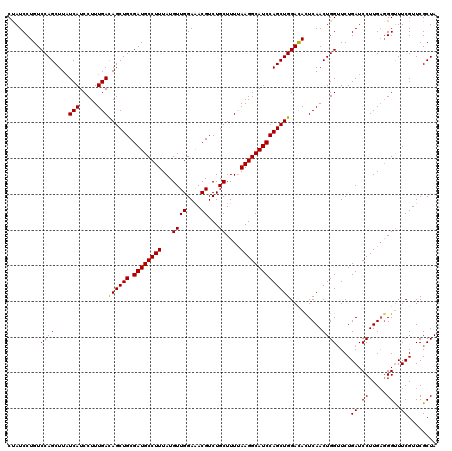
| Location | 3,717,421 – 3,717,541 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -33.29 |
| Energy contribution | -33.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3717421 120 - 22407834 UAGCGAACGAAGCCCGCAAGGAUCAGAUCCAGUUGAAUAUCCAGCUGGAUGCCUUGAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAGAAGCUGGACAGGAUGG ..((.......))((....))(((...(((((((...(((((((((((((((((((.((((....)))).))......))))))).)))))......)))))...)))))))...))).. ( -40.20) >DroSec_CAF1 179739 120 - 1 UAGCGAACGAAACCGUCAAGGAUCAGAACCAGUUGAGUGUCCAGCUGGAUGCCUUAAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUAUCAAAGGAUGAUAAGCUGGACAGGAUAG ............((.....))(((....((((((...((((((((((((((((((.......((....)).......)))))))).)))))......)))))...))))))....))).. ( -33.64) >DroSim_CAF1 188618 120 - 1 UAGCGAACGAAACCCUCAAGGAUCAGAACCAGUUGAGUGUCCAGCUGGAUGCCUUAAAAGCAGACGUCUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAUAAGCUGGACAGGAUAG ............(((((((((.......))..)))))(((((((((.((((((((.......((....)).......)))))))).....(((((.....)))))))))))))))).... ( -33.44) >consensus UAGCGAACGAAACCCUCAAGGAUCAGAACCAGUUGAGUGUCCAGCUGGAUGCCUUAAAAGCAGACGUUUCCAACAUAAAGGCAUCGCAGCUGUCAAAGGAUGAUAAGCUGGACAGGAUAG ............((.....))(((....((((((...((((((((((((((((((.......((....)).......)))))))).)))))......)))))...))))))....))).. (-33.29 = -33.07 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:42 2006