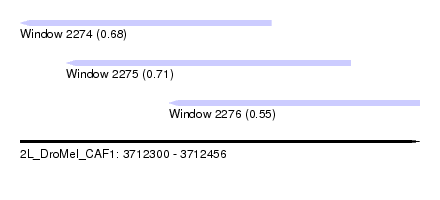
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,712,300 – 3,712,456 |
| Length | 156 |
| Max. P | 0.706803 |

| Location | 3,712,300 – 3,712,398 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.18 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3712300 98 - 22407834 ACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAAGAUCAAAGGACUUCUAGAUCCGGAAACCUGCAUCAAGUGUCCGCGGUGUCAUAAGUCCAUUU---------------------- ...(....)...((((((((....))))))))...........((((((..(.(((((((...((......))..)))).))).)...))))))....---------------------- ( -29.90) >DroSec_CAF1 174651 98 - 1 ACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCGCAGGUCUUCUAGAUCCGGAAACUUGCAUCAAGUGUCCGCGGUGUCAUAAGUCCAUUU---------------------- ....((((....((((((((....))))))))......)))).((.(((..(.(((((((.(((((...))))).)))).))).)...))).))....---------------------- ( -31.70) >DroSim_CAF1 183542 98 - 1 ACGCGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCGUAGGUCUUCUAGAUCCGGAAACUUGCAUCAAGUGUCCGCGGUGUCAUAAGUCCAUUU---------------------- .(((........((((((((....))))))))...(((((.(((.....))))))))(((.(((((...))))).)))))).................---------------------- ( -33.10) >DroEre_CAF1 187820 98 - 1 CAACCUGAUAACGCAGAUACAUCCGUGUCUGCACAGGGUCACAGGUAAUCGAGAUCCGGAAACCUGCCUCAAGUGCCCGCGGUGUCAAAAGCCCAUUU---------------------- .....(((((.(((((((((....))))))((((.(((...(((((..(((.....)))..))))))))...))))..))).)))))...........---------------------- ( -28.90) >DroYak_CAF1 188212 98 - 1 AAAACUGAGAACGCAGAUACAUCCGUGUUUGCACAGGAUUAAAGGACUUUUGGAUCCGGACACCUGCCCCAAUUGUCCGCGGUGUCAAAAGCCCAUUU---------------------- ............((((((((....))))))))...........((.((((((((((((((((..((...))..)))))).))).))))))).))....---------------------- ( -29.50) >DroAna_CAF1 199115 120 - 1 UACUGUGAUUUCGGUGAUAUAUCCCUGUCUGCACAGCGUUAAAGGUCGUGUUGAUCCGGACACGGAUUCGAAGUGUCCUCUUUGUAAAAAACCUAUUCACAAAGUGUUUACAAAGGCUAG ..(((((....(((.((....)).)))....)))))......(((.((..((((((((....)))).))))..)).)))(((((((((....((........))..)))))))))..... ( -29.90) >consensus AAACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCAAAGGUCUUCUAGAUCCGGAAACCUGCAUCAAGUGUCCGCGGUGUCAAAAGCCCAUUU______________________ .....(((.(.(((((((((....))))))((((.(((((............)))))(....).........))))..))).).)))................................. (-15.53 = -15.87 + 0.34)

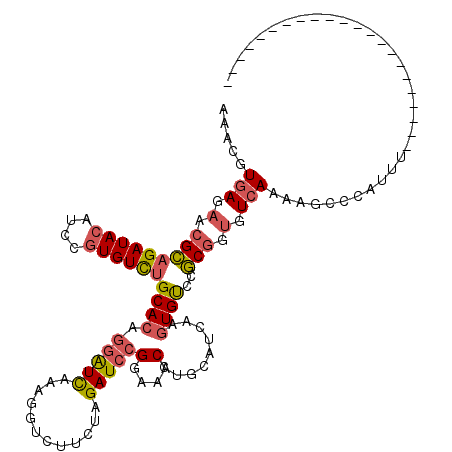
| Location | 3,712,318 – 3,712,429 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3712318 111 - 22407834 ---------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGAACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAAGAUCAAAGGACUUCUAGAUCCGGAAACCUGCAUCAAGUGUCCGC ---------.(((((((...((.....))((((....(((.(.(....)...((((((((....))))))))....................).)))(....)))))....))))))).. ( -31.70) >DroSec_CAF1 174669 111 - 1 ---------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGCACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCGCAGGUCUUCUAGAUCCGGAAACUUGCAUCAAGUGUCCGC ---------.((((((((((..((........))...)))....(..((...((((((((....))))))))...(((((..((.....)).))))).....))..)....))))))).. ( -38.60) >DroSim_CAF1 183560 111 - 1 ---------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGCAACGCGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCGUAGGUCUUCUAGAUCCGGAAACUUGCAUCAAGUGUCCGC ---------.(((((((...((.((..((((.((........))))))....((((((((....))))))))...)).))((((((.((((......))))))))))....))))))).. ( -36.70) >DroEre_CAF1 187838 111 - 1 ---------UGGAUAUUGCAGAUCCAAACACAGGCAAUAACAACCUGAUAACGCAGAUACAUCCGUGUCUGCACAGGGUCACAGGUAAUCGAGAUCCGGAAACCUGCCUCAAGUGCCCGC ---------.((.((((...(((((.....((((.........)))).....((((((((....))))))))...))))).(((((..(((.....)))..))))).....)))).)).. ( -34.50) >DroYak_CAF1 188230 111 - 1 ---------UGGACAUUGAAGAGACAAUCGCAGGAAAGGAAAAACUGAGAACGCAGAUACAUCCGUGUUUGCACAGGAUUAAAGGACUUUUGGAUCCGGACACCUGCCCCAAUUGUCCGC ---------.((((((((..((.....))(((((..................((((((((....))))))))...(((((.(((....))).))))).....)))))..))).))))).. ( -31.70) >DroAna_CAF1 199155 120 - 1 AAAACAUUACUGAUAUUGUCGAAAAAAGAAGAAAAAAAGAUACUGUGAUUUCGGUGAUAUAUCCCUGUCUGCACAGCGUUAAAGGUCGUGUUGAUCCGGACACGGAUUCGAAGUGUCCUC ...........((((((.(((((...............((..(((((....(((.((....)).)))....)))))..)).....(((((((......))))))).)))))))))))... ( -26.30) >consensus _________UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGAAAACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUCAAAGGUCUUCUAGAUCCGGAAACCUGCAUCAAGUGUCCGC ..........(((((((...................................((((((((....))))))))...(((((............)))))..............))))))).. (-19.20 = -19.82 + 0.61)

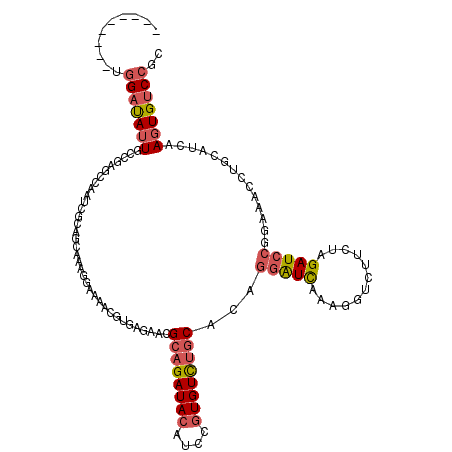
| Location | 3,712,358 – 3,712,456 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.54 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -9.82 |
| Energy contribution | -10.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3712358 98 - 22407834 ---CUUU---------AUUUCAGAUACUCCACCAGCGGU----------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGAACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAAGAUC ---....---------.(((((.....(((....(((((----------(((...........))))))))......))).....)))))..((((((((....))))))))........ ( -27.50) >DroSec_CAF1 174709 98 - 1 ---CUUU---------AUUUUAGAUACUGCACCAGCGGU----------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGCACACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUC ---....---------......(((.((((((..(((((----------(((...........)))))))).((....))....))).....((((((((....)))))))).))).))) ( -30.40) >DroSim_CAF1 183600 98 - 1 ---CUUU---------AUUUUAGAUACUCCACCAGCGGU----------UGGAUAUUGCCGAGCCAAUCGCAGCAAAGCAACGCGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUC ---....---------...........(((....(((((----------(((...........))))))))......((...((((....))))((((((....))))))))...))).. ( -28.50) >DroEre_CAF1 187878 101 - 1 AACAAAU---------CUUUCAGAUAAACCGCCAACAAC----------UGGAUAUUGCAGAUCCAAACACAGGCAAUAACAACCUGAUAACGCAGAUACAUCCGUGUCUGCACAGGGUC .....((---------(.....))).....(((......----------(((((.......)))))......)))........((((.....((((((((....)))))))).))))... ( -24.80) >DroYak_CAF1 188270 101 - 1 AACAACU---------AUUUCAGAUACUCCACCCGCGAU----------UGGACAUUGAAGAGACAAUCGCAGGAAAGGAAAAACUGAGAACGCAGAUACAUCCGUGUUUGCACAGGAUU .......---------.((((((....(((.((.(((((----------((.............))))))).))...)))....))))))..((((((((....))))))))........ ( -29.82) >DroAna_CAF1 199195 116 - 1 ---ACUAAACAAUACAUUUUUAGAUACUCA-GAAGAAAUCAAAACAUUACUGAUAUUGUCGAAAAAAGAAGAAAAAAAGAUACUGUGAUUUCGGUGAUAUAUCCCUGUCUGCACAGCGUU ---.............(((((.((((.(((-(.................))))...)))).)))))............((..(((((....(((.((....)).)))....)))))..)) ( -14.13) >consensus ___AUUU_________AUUUCAGAUACUCCACCAGCGAU__________UGGAUAUUGCCGAGCCAAUCGCAGCAAAGGAAAACGUGAGAACGCAGAUACAUCCGUGUCUGCACAGGAUC ............................................................................................((((((((....))))))))........ ( -9.82 = -10.18 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:35 2006