| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,708,011 – 3,708,118 |
| Length | 107 |
| Max. P | 0.703457 |

| Location | 3,708,011 – 3,708,118 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.85 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
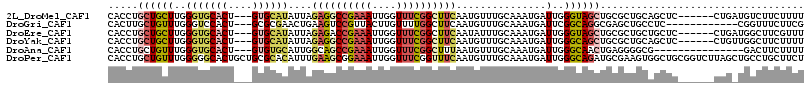
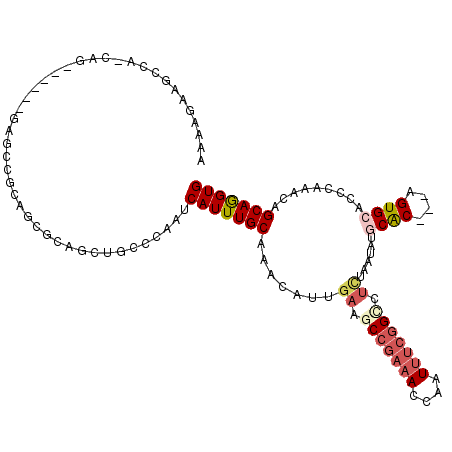
>2L_DroMel_CAF1 3708011 107 + 22407834 CACCUGCUGCUUGGGUGCACU---GUGCAUAUUAGAGGCCGAAAUUGGUUUCGGCUUCAAUGUUUGCAAAUGAUUGGGUAGCUGCGCUGCAGCUC------CUGAUGUCUUCUUUU (((((.......)))))((..---.((((.((..((((((((((....))))))))))...)).))))..))((..((.((((((...)))))))------)..)).......... ( -38.80) >DroGri_CAF1 214910 101 + 1 CACUUGCUGUUUGGGUCCACU---GCGCGAACUGAAGUCCGUUACUUGUUUUGGCUUCAAUGUUUGCAAAUGAUUCGGCAGGCGAGCUGCCUC------------CGGUUUCUUCG ((.((((.(((((.((.....---)).)))))(((((.(((..(....)..))))))))......)))).))..((((.((((.....)))))------------)))........ ( -27.80) >DroEre_CAF1 183684 107 + 1 CACCUGCUGCUUGGGUGCACU---GUGCAUAUUAGAGACCGAAAUUGGUUUCGGCUUCAAUAUUUGCAAAUGAUUGGGUAGCUGCGCUGCUGCUC------CUGAUGGCUUCGUUU .....((((((..(...((..---.((((((((..((.((((((....))))))))..))))..))))..)).)..)))))).(((..(((((..------..).))))..))).. ( -31.90) >DroYak_CAF1 184043 107 + 1 CACCUGCUGCUUGGGUGCACU---GUGCAUAUUAGAGGCCGAAAUUGGUUUCGGCUUCAAUGUUUGCAAAUGAUUGGGCAGCUGCGCUGCAGCUC------CUGUUGGCUUCUUUU .....(((((...((((((((---((.((.((((((((((((((....))))))))))..((....))..)))))).)))).))))))))))).(------(....))........ ( -39.50) >DroAna_CAF1 193730 98 + 1 CACCUGCUGUUUGGGUGCACU---GUGUGCAUUGGCAGCCGAAAUUGGUUUCGGCUUUAAUGUUUGCAAAUGAUUGGGCAACUGAGGGGCG---------------GACUUCUUUU ...(((((.((..(.(((..(---(((.(((((((.((((((((....))))))))))))))).)))).........))).)..)).))))---------------)......... ( -32.70) >DroPer_CAF1 195633 116 + 1 CACCUGCUGUUUGGGGGCACUGCUGCGCACAUUUGAAGCGGAAAUUGGUUUCGGUUUCAAUGUUUGCAAAUGAUUGGGCAGAUGCGAAGUGGCUGCGGUCUUAGCUGCCUGCUUCU ..(((.......))).(((((((((((.(((((.(((((.((((....)))).)))))))))).))).........))))).)))((((((((.((.......)).))).))))). ( -39.70) >consensus CACCUGCUGCUUGGGUGCACU___GUGCAUAUUAGAGGCCGAAAUUGGUUUCGGCUUCAAUGUUUGCAAAUGAUUGGGCAGCUGCGCUGCAGCUC______CUG_UGGCUUCUUUU .....((((((..(((((((....))))))....((((((((((....))))))))))...............)..)))))).................................. (-20.82 = -21.85 + 1.03)
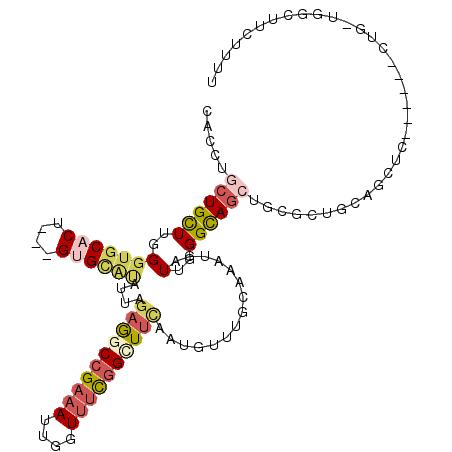
| Location | 3,708,011 – 3,708,118 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.35 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3708011 107 - 22407834 AAAAGAAGACAUCAG------GAGCUGCAGCGCAGCUACCCAAUCAUUUGCAAACAUUGAAGCCGAAACCAAUUUCGGCCUCUAAUAUGCAC---AGUGCACCCAAGCAGCAGGUG ..............(------(((((((...)))))).))....(((((((...(((.((.(((((((....))))))).))....)))...---..(((......)))))))))) ( -30.50) >DroGri_CAF1 214910 101 - 1 CGAAGAAACCG------------GAGGCAGCUCGCCUGCCGAAUCAUUUGCAAACAUUGAAGCCAAAACAAGUAACGGACUUCAGUUCGCGC---AGUGGACCCAAACAGCAAGUG .........((------------(((((.....)))).)))...(((((((......(((((((............)).)))))((((((..---.)))))).......))))))) ( -28.20) >DroEre_CAF1 183684 107 - 1 AAACGAAGCCAUCAG------GAGCAGCAGCGCAGCUACCCAAUCAUUUGCAAAUAUUGAAGCCGAAACCAAUUUCGGUCUCUAAUAUGCAC---AGUGCACCCAAGCAGCAGGUG ..............(------(.(.(((......))).)))...(((((((..(((((((..((((((....))))))..)).)))))....---..(((......)))))))))) ( -26.00) >DroYak_CAF1 184043 107 - 1 AAAAGAAGCCAACAG------GAGCUGCAGCGCAGCUGCCCAAUCAUUUGCAAACAUUGAAGCCGAAACCAAUUUCGGCCUCUAAUAUGCAC---AGUGCACCCAAGCAGCAGGUG ..............(------(((((((...)))))).))....(((((((...(((.((.(((((((....))))))).))....)))...---..(((......)))))))))) ( -30.50) >DroAna_CAF1 193730 98 - 1 AAAAGAAGUC---------------CGCCCCUCAGUUGCCCAAUCAUUUGCAAACAUUAAAGCCGAAACCAAUUUCGGCUGCCAAUGCACAC---AGUGCACCCAAACAGCAGGUG ..........---------------...................(((((((.........((((((((....)))))))).....(((((..---.)))))........))))))) ( -22.30) >DroPer_CAF1 195633 116 - 1 AGAAGCAGGCAGCUAAGACCGCAGCCACUUCGCAUCUGCCCAAUCAUUUGCAAACAUUGAAACCGAAACCAAUUUCCGCUUCAAAUGUGCGCAGCAGUGCCCCCAAACAGCAGGUG .((((..(((.((.......)).))).)))).(((((((........((((..((((((((.(.((((....)))).).))).)))))..))))..((........)).))))))) ( -28.90) >consensus AAAAGAAGCCA_CAG______GAGCCGCAGCGCAGCUGCCCAAUCAUUUGCAAACAUUGAAGCCGAAACCAAUUUCGGCCUCUAAUAUGCAC___AGUGCACCCAAACAGCAGGUG ............................................(((((((.......((.(((((((....))))))).))......((((....)))).........))))))) (-14.24 = -15.35 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:31 2006