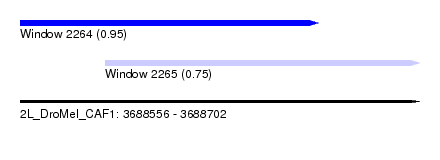
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,688,556 – 3,688,702 |
| Length | 146 |
| Max. P | 0.946673 |

| Location | 3,688,556 – 3,688,665 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.52 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
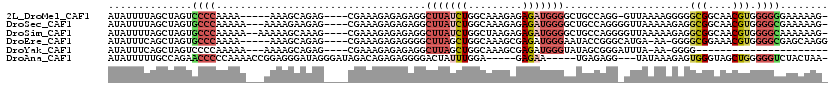
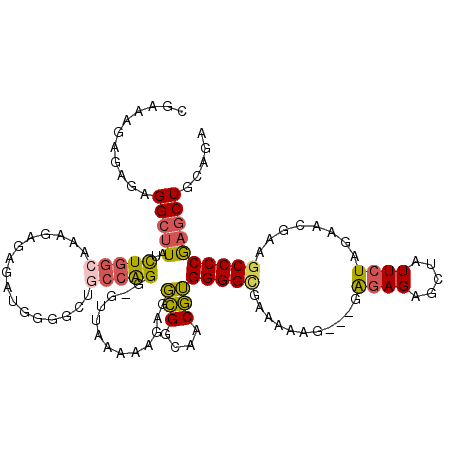
>2L_DroMel_CAF1 3688556 109 + 22407834 AUAUUUUAGCUAGUCCCCAAAA-----AAAGCAGAG----CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGG-GUUAAAAGGGGGCGGCAACGUGGGGGGAAAAAG- ........(((.((((((....-----.((((....----(....)......)))).(((((((..............)))))))-.......))))))))).................- ( -30.14) >DroSec_CAF1 150900 112 + 1 AUAUUUUAGCUAGUGCCCAAAAA---AAAAGAAGAG----CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGGGGUUAAAAAGAGGCGGCAACGUGGGGCGAAAAAG- .............(((((.....---...(((.(((----(...........)))).)))...........(((..((((((..............))))))..))).)))))......- ( -27.34) >DroSim_CAF1 159921 113 + 1 AUAUUUUAGCUAGUGCCCAAAAA--AAAAAGCAAAG----CGAAAGAGAGAGGCUUAUCUGGCUAAGAGAGAUGGGGCUGCCAGGGGUUAAAAAGAGGCGGCAACGUGGGGCAAAAAAG- .............(((((.....--...((((....----(....)......)))).(((.....)))...(((..((((((..............))))))..))).)))))......- ( -28.64) >DroEre_CAF1 155945 109 + 1 AUAUUUCAGCUAGUGCCCAAAA-----AAAGCAGAG----CGAAAGAGAGGGGCUUAGCUGGCAAAGCGAGAUGGGAAUACCGGGCAUGA-AA-GGGGCGGAAACGUGGGGCGAGCAAGG ........(((.((((((....-----.((((....----(....)......)))).(((.....))).....((.....))))))))..-..-...(((....)))..)))........ ( -29.90) >DroYak_CAF1 161353 89 + 1 AUAUUUCAGCUAGUCCCCAAAAA---AAAAGCAGAG----CGAAAGAGAGAGGCUUAGCUGGCAAAGCGAGAUGGGUAUAGCGGGAUUUA-AA-GGGG---------------------- .((..((.((((...((((....---..((((....----(....)......)))).(((.....)))....))))..))))..))..))-..-....---------------------- ( -21.30) >DroAna_CAF1 167514 106 + 1 AUAUUUUUGCCAGAACCCCCAAAACCGGAGGGAUAGGGAUAGACAGAGAGGGGACUAUUUGGA-----GAGAA-----UGAGAGG---UAUAAAGAGUGGGUAGCUGGGGGUCUACUAA- .((((((..(((((.(((((......)).)))................((....)).))))).-----.))))-----)).....---...........((((((.....).)))))..- ( -22.80) >consensus AUAUUUUAGCUAGUGCCCAAAAA___AAAAGCAGAG____CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGG_GUUAAAAAGAGGCGGCAACGUGGGGCGAAAAAG_ ..............((((...................................(((((((.........))))))).....................(((....))).))))........ ( -8.82 = -9.77 + 0.95)
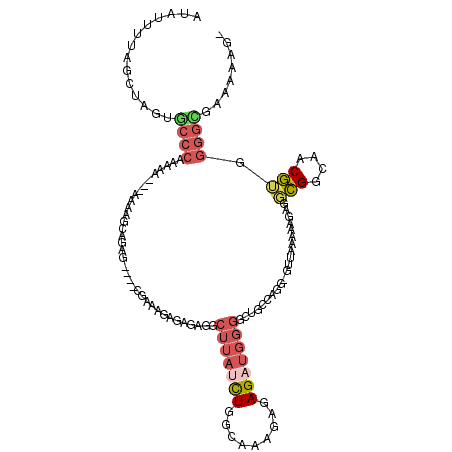
| Location | 3,688,587 – 3,688,702 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.64 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3688587 115 + 22407834 CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGG-GUUAAAAGGGGGCGGCAACGUGGGGGGAAAAAG---GACAGAGCUAUUCUAGAACGAAGCCCCGAGCUUCAGA (....)...(((((((.(((((((..............)))))))-........((((((....)((....((((...(---(......)).))))...))...))))))))))))... ( -34.34) >DroSec_CAF1 150933 116 + 1 CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGGGGUUAAAAAGAGGCGGCAACGUGGGGCGAAAAAG---GAGAGAGCUAUUCUAGAACGAAGCCCCGAGCUGCAGA (....)...(.(((((.(((.....)))...(((..((((((..............))))))..)))(((((.......---.((((.....))))........)))))))))).)... ( -32.83) >DroSim_CAF1 159955 116 + 1 CGAAAGAGAGAGGCUUAUCUGGCUAAGAGAGAUGGGGCUGCCAGGGGUUAAAAAGAGGCGGCAACGUGGGGCAAAAAAG---GAGAGAGCUAUUCUAGAACGAAGCCCCGAGCUGCAGA (....)...(.(((((.(((.....)))...(((..((((((..............))))))..)))(((((.......---.((((.....))))........)))))))))).)... ( -31.73) >DroEre_CAF1 155976 117 + 1 CGAAAGAGAGGGGCUUAGCUGGCAAAGCGAGAUGGGAAUACCGGGCAUGA-AA-GGGGCGGAAACGUGGGGCGAGCAAGGGAGAGAGAGCUAUUCUAGAGCGAAGCCCCAAGCUUCUGA (....)..((((((((.(((.....)))(...(((.....)))..)....-..-...(((....)))(((((..((...((((((....)).))))...))...))))))))))))).. ( -37.30) >DroAna_CAF1 167554 98 + 1 AGACAGAGAGGGGACUAUUUGGA-----GAGAA-----UGAGAGG---UAUAAAGAGUGGGUAGCUGGGGGUCUACUAA---GGGAGGAUGGUUCUAGAACGCCGCCCCAAGCU----- ...((((.((....)).))))..-----.....-----.......---..............((((((((((((.(...---..).))))(((........))).)))).))))----- ( -22.00) >consensus CGAAAGAGAGAGGCUUAUCUGGCAAAGAGAGAUGGGGCUGCCAGG_GUUAAAAAGAGGCGGCAACGUGGGGCGAAAAAG___GAGAGAGCUAUUCUAGAACGAAGCCCCGAGCUGCAGA ...........(((((..(((((................))))).............(((....)))(((((...........((((.....))))........))))))))))..... (-18.62 = -18.74 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:20 2006