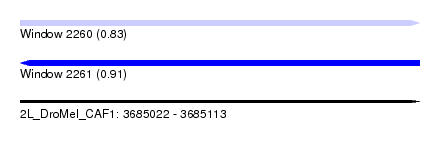
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,685,022 – 3,685,113 |
| Length | 91 |
| Max. P | 0.908540 |

| Location | 3,685,022 – 3,685,113 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -27.05 |
| Energy contribution | -26.47 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
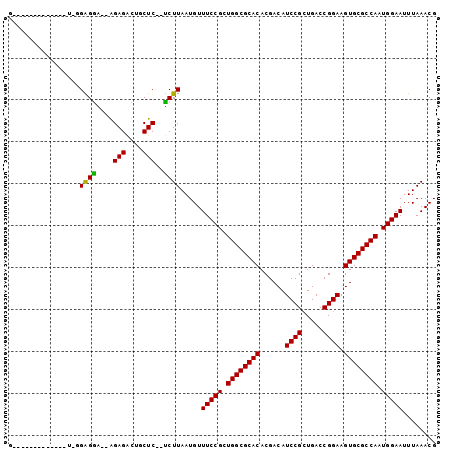
>2L_DroMel_CAF1 3685022 91 + 22407834 GG---UG----CACUCGGAGGA--GGAGACUGCUC--UCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG ..---..----.....((((.(--(....)).)))--).((((...(((((.((((((((......((((.....)))).)))))))).))))).))))... ( -33.20) >DroPse_CAF1 172844 84 + 1 G----------------GAGAACGAGAGGUUGCUC--UCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG .----------------......(((((....)))--))((((...(((((.((((((((......((((.....)))).)))))))).))))).))))... ( -30.10) >DroEre_CAF1 152399 86 + 1 --------------UUGGGGGA--GGAGACUGCUCUCUCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG --------------..((((((--(....)).)))))..((((...(((((.((((((((......((((.....)))).)))))))).))))).))))... ( -32.70) >DroYak_CAF1 157856 95 + 1 GG---GAGGCUUAUUUGGAGGA--GGAGACUGCUC--UCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG .(---((((((((...((((.(--(....)).)))--)..))).))))))..((((((((......((((.....)))).)))))))).............. ( -35.10) >DroAna_CAF1 163498 94 + 1 AGUCAGG----GAAACAGGGGU--AGAGAUUGCUC--CCUUAAUGUUUCCGCUGGCGCACACGACAUCCGGUGACCGGAAGUGCGCCAAUGGAAUUUAAACG .((((((----((((((((((.--((......)))--)))...))))))).)))))((.(((....(((((...))))).))).))................ ( -34.50) >DroPer_CAF1 172861 84 + 1 G----------------GAGAACGAGAGGUUGCUC--UCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG .----------------......(((((....)))--))((((...(((((.((((((((......((((.....)))).)))))))).))))).))))... ( -30.10) >consensus G_____________U_GGAGGA__AGAGACUGCUC__UCUUAAUGUUUCCGCUGGCGCACACGACAUCCGCUGACCGGAAGUGCGCCAAUGGAAUUUAAACG .................((((....(((....)))..)))).....(((((.((((((((......((((.....)))).)))))))).)))))........ (-27.05 = -26.47 + -0.58)


| Location | 3,685,022 – 3,685,113 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
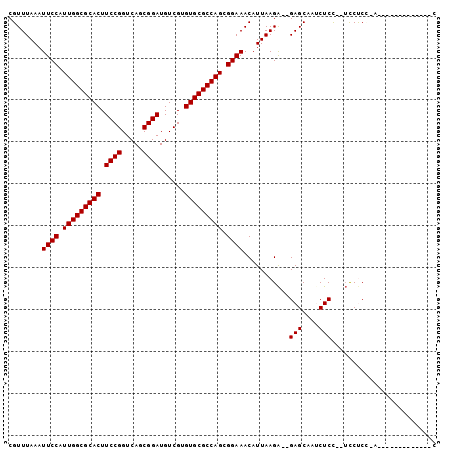
>2L_DroMel_CAF1 3685022 91 - 22407834 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGA--GAGCAGUCUCC--UCCUCCGAGUG----CA---CC ........((((.(((((((((.((((.....))))......))))))))).)))).........--..(((.((...--......)).))----).---.. ( -30.70) >DroPse_CAF1 172844 84 - 1 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGA--GAGCAACCUCUCGUUCUC----------------C ........((((.(((((((((.((((.....))))......))))))))).)))).......((--((((........))))))----------------. ( -32.40) >DroEre_CAF1 152399 86 - 1 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGAGAGAGCAGUCUCC--UCCCCCAA-------------- ........((((.(((((((((.((((.....))))......))))))))).)))).......(((.(((....))))--))......-------------- ( -29.90) >DroYak_CAF1 157856 95 - 1 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGA--GAGCAGUCUCC--UCCUCCAAAUAAGCCUC---CC .(((((..((((.(((((((((.((((.....))))......))))))))).)))).......((--((....)))).--.........)))))...---.. ( -30.40) >DroAna_CAF1 163498 94 - 1 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCACCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGG--GAGCAAUCUCU--ACCCCUGUUUC----CCUGACU .(((.........(((((((((.(((((...)))))......))))))))).(((((((...(((--((....)))))--.....))))))----)..))). ( -35.00) >DroPer_CAF1 172861 84 - 1 CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGA--GAGCAACCUCUCGUUCUC----------------C ........((((.(((((((((.((((.....))))......))))))))).)))).......((--((((........))))))----------------. ( -32.40) >consensus CGUUUAAAUUCCAUUGGCGCACUUCCGGUCAGCGGAUGUCGUGUGCGCCAGCGGAAACAUUAAGA__GAGCAAUCUCC__UCCUCC_A_____________C ........((((.(((((((((.((((.....))))......))))))))).))))...........(((....)))......................... (-26.25 = -26.25 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:15 2006