
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,684,825 – 3,684,916 |
| Length | 91 |
| Max. P | 0.825263 |

| Location | 3,684,825 – 3,684,916 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.68 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.49 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
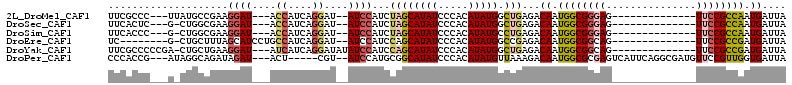
>2L_DroMel_CAF1 3684825 91 + 22407834 UUCGCCC---UUAUGCCGAAGGAU---ACCAUCAGGAU--AUCCAUCUAGCAUAUCCCACAUAUGGCUGAGACAAUGGCGGGAG--------------UUCCGCCAAUGAUUA ((((((.---.(((((.((.((((---(((....)).)--)))).))..)))))..........))).))).((.((((((...--------------..)))))).)).... ( -27.40) >DroSec_CAF1 147212 90 + 1 UUCACUC---G-CUGGCGAAGGAU---ACCAUCAGGAU--AUCCAUCUAGCAUAUCCCACAUAUGGCUGAGACAAUGGCGGGAG--------------UUCCGCCAAUGAUUA ....(((---(-((((....((((---(((....)).)--))))..))))).....(((....)))..))).((.((((((...--------------..)))))).)).... ( -29.20) >DroSim_CAF1 156286 90 + 1 UUCACCC---G-CUGGCGAAGGAU---ACCAUCAGGAU--AUCCAUCUAGCAUAUCCCACAUAUGCCUGAGACAAUGGCGGGAG--------------UUCCGCCAAUGAUUA (((.((.---.-..)).)))((((---(((....)).)--)))).((..((((((.....))))))..))..((.((((((...--------------..)))))).)).... ( -28.40) >DroEre_CAF1 152217 88 + 1 UC--------G-CUGCUUUAGCAUCCUGCCAUCAGGAU--AUCCAUCCAGCAUAUCCCACAUAUGGCCGAGACAAUGGCGGCAG--------------UUCCGCCGAUGAUUA ..--------(-(((...)))).(((.(((((..((((--((.(.....).)))))).....))))).).))((.((((((...--------------..)))))).)).... ( -24.30) >DroYak_CAF1 157655 95 + 1 UUCGCCCCCGA-CUGCUGAAGGAU---AUCAUCAGGAUAUAUCCAUCCAGCAUAUCCCACAUAUGGCUGAGACAAUGGCGGCAG--------------UUCCGCCGAUGAUUA .......((..-(....)..))..---((((((.(((....))).((((((((((.....)))).)))).))....(((((...--------------..))))))))))).. ( -24.60) >DroPer_CAF1 172669 100 + 1 CCCACCG---AUAGGCAGAUAGAU---ACU-----CGU--AUCCAUGCGGCAUAUCCCACAUAUGUUAAAGACAAUGGCGCGAGUCAUUCAGGCGAUGUUCCGUUGGUGAUUA ..(((((---((..(((....(((---(..-----..)--)))..)))((.(((((((.....((((...))))(((((....)))))...)).))))).))))))))).... ( -30.70) >consensus UUCACCC___G_CUGCCGAAGGAU___ACCAUCAGGAU__AUCCAUCCAGCAUAUCCCACAUAUGGCUGAGACAAUGGCGGGAG______________UUCCGCCAAUGAUUA ....................((((....((....))....))))...((((((((.....))))).)))...((.((((((((...............)))))))).)).... (-13.77 = -14.49 + 0.72)
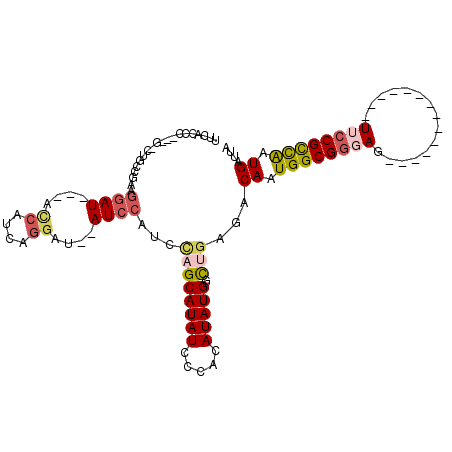

| Location | 3,684,825 – 3,684,916 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.68 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -13.91 |
| Energy contribution | -15.36 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
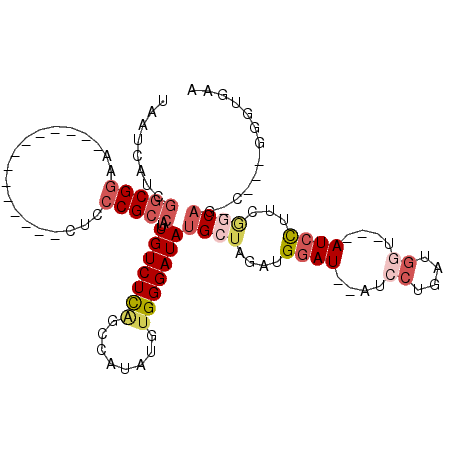
>2L_DroMel_CAF1 3684825 91 - 22407834 UAAUCAUUGGCGGAA--------------CUCCCGCCAUUGUCUCAGCCAUAUGUGGGAUAUGCUAGAUGGAU--AUCCUGAUGGU---AUCCUUCGGCAUAA---GGGCGAA .......((((((..--------------...))))))((((((...(((....)))..((((((.((.((((--(((.....)))---)))).)))))))).---)))))). ( -33.00) >DroSec_CAF1 147212 90 - 1 UAAUCAUUGGCGGAA--------------CUCCCGCCAUUGUCUCAGCCAUAUGUGGGAUAUGCUAGAUGGAU--AUCCUGAUGGU---AUCCUUCGCCAG-C---GAGUGAA ...((((((((((..--------------...)))))..(((((((........)))))))((((.((.((((--(((.....)))---)))).))...))-)---)))))). ( -30.80) >DroSim_CAF1 156286 90 - 1 UAAUCAUUGGCGGAA--------------CUCCCGCCAUUGUCUCAGGCAUAUGUGGGAUAUGCUAGAUGGAU--AUCCUGAUGGU---AUCCUUCGCCAG-C---GGGUGAA ....((.((((((..--------------...)))))).))..((.(((((((.....))))))).)).((((--(((.....)))---)))).(((((..-.---.))))). ( -33.00) >DroEre_CAF1 152217 88 - 1 UAAUCAUCGGCGGAA--------------CUGCCGCCAUUGUCUCGGCCAUAUGUGGGAUAUGCUGGAUGGAU--AUCCUGAUGGCAGGAUGCUAAAGCAG-C--------GA ........(((((..--------------...)))))..(((((((........)))))))(((((..(((.(--((((((....))))))))))...)))-)--------). ( -31.00) >DroYak_CAF1 157655 95 - 1 UAAUCAUCGGCGGAA--------------CUGCCGCCAUUGUCUCAGCCAUAUGUGGGAUAUGCUGGAUGGAUAUAUCCUGAUGAU---AUCCUUCAGCAG-UCGGGGGCGAA ........(((((..--------------...))))).(((((((..(((....)))(((.(((((((.((((((.........))---))))))))))))-)).))))))). ( -36.50) >DroPer_CAF1 172669 100 - 1 UAAUCACCAACGGAACAUCGCCUGAAUGACUCGCGCCAUUGUCUUUAACAUAUGUGGGAUAUGCCGCAUGGAU--ACG-----AGU---AUCUAUCUGCCUAU---CGGUGGG ...(((((..(((........)))....(((((..((..(((.....))).((((((......))))))))..--.))-----)))---..............---.))))). ( -23.00) >consensus UAAUCAUCGGCGGAA______________CUCCCGCCAUUGUCUCAGCCAUAUGUGGGAUAUGCUAGAUGGAU__AUCCUGAUGGU___AUCCUUCGGCAG_C___GGGUGAA ........(((((...................)))))..(((((((........)))))))((((....((((....((....))....))))...))))............. (-13.91 = -15.36 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:13 2006